Origin and evolution of the chicken leukocyte receptor complex
- PMID: 15753291
- PMCID: PMC554838
- DOI: 10.1073/pnas.0501040102
Origin and evolution of the chicken leukocyte receptor complex
Abstract
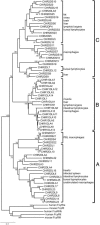
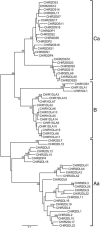
In mammals, the cell surface receptors encoded by the leukocyte receptor complex (LRC) regulate the activity of T lymphocytes and B lymphocytes, as well as that of natural killer cells, and thus provide protection against pathogens and parasites. The chicken genome encodes many Ig-like receptors that are homologous to the LRC receptors. The chicken Ig-like receptor (CHIR) genes are members of a large monophyletic gene family and are organized into genomic clusters, which are in conserved synteny with the mammalian LRC. One-third of CHIR genes encode polypeptide molecules that contain both activating and inhibitory motifs. These genes are present in different phylogenetic groups, suggesting that the primordial CHIR gene could have encoded both types of motifs in a single molecule. In contrast to the mammalian LRC genes, the CHIR genes with similar function (inhibition or activation) are evolutionarily closely related. We propose that, in addition to recombination, single nucleotide substitutions played an important role in the generation of receptors with different functions. Structural models and amino acid analyses of the CHIR proteins reveal the presence of different types of Ig-like domains in the same phylogenetic groups, as well as sharing of conserved residues and conserved changes of residues between different CHIR groups and between CHIRs and LRCs. Our data support the notion that the CHIR gene clusters are regions homologous to the mammalian LRC gene cluster and favor a model of evolution by repeated processes of birth and death (expansion-contraction) of the Ig-like receptor genes.
Figures


References
-
- Lanier, L. L. (2003) Curr. Opin. Immunol. 15, 308–314. - PubMed
-
- Martin, A. M., Kulski, J. K., Witt, C., Pontarotti, P. & Christiansen, F. T. (2002) Trends Immunol. 23, 81–88. - PubMed
-
- Trowsdale, J., Barten, R., Haude, A., Stewart, C. A., Beck, S. & Wilson, M. J. (2001) Immunol. Rev. 181, 20–38. - PubMed
-
- Volz, A., Wende, H., Laun, K. & Ziegler, A. (2001) Immunol. Rev. 181, 39–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

