Secondary structure in the target as a confounding factor in synthetic oligomer microarray design
- PMID: 15755320
- PMCID: PMC555549
- DOI: 10.1186/1471-2164-6-31
Secondary structure in the target as a confounding factor in synthetic oligomer microarray design
Abstract
Background: Secondary structure in the target is a property not usually considered in software applications for design of optimal custom oligonucleotide probes. It is frequently assumed that eliminating self-complementarity, or screening for secondary structure in the probe, is sufficient to avoid interference with hybridization by stable secondary structures in the probe binding site. Prediction and thermodynamic analysis of secondary structure formation in a genome-wide set of transcripts from Brucella suis 1330 demonstrates that the properties of the target molecule have the potential to strongly influence the rate and extent of hybridization between transcript and tethered oligonucleotide probe in a microarray experiment.
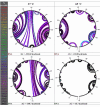
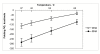
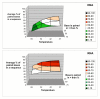
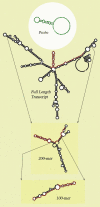
Results: Despite the relatively high hybridization temperatures and 1M monovalent salt imposed in the modeling process to approximate hybridization conditions used in the laboratory, we find that parts of the target molecules are likely to be inaccessible to intermolecular hybridization due to the formation of stable intramolecular secondary structure. For example, at 65 degrees C, 28 +/- 7% of the average cDNA target sequence is predicted to be inaccessible to hybridization. We also analyzed the specific binding sites of a set of 70mer probes previously designed for Brucella using a freely available oligo design software package. 21 +/- 13% of the nucleotides in each probe binding site are within a double-stranded structure in over half of the folds predicted for the cDNA target at 65 degrees C. The intramolecular structures formed are more stable and extensive when an RNA target is modeled rather than cDNA. When random shearing of the target is modeled for fragments of 200, 100 and 50 nt, an overall destabilization of secondary structure is predicted, but shearing does not eliminate secondary structure.
Conclusion: Secondary structure in the target is pervasive, and a significant fraction of the target is found in double stranded conformations even at high temperature. Stable structure in the target has the potential to interfere with hybridization and should be a factor in interpretation of microarray results, as well as an explicit criterion in array design. Inclusion of this property in an oligonucleotide design procedure would change the definition of an optimal oligonucleotide significantly.
Figures


Similar articles
-
A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus.BMC Genomics. 2005 Jun 17;6:95. doi: 10.1186/1471-2164-6-95. BMC Genomics. 2005. PMID: 15963225 Free PMC article.
-
Analysis of crucial factors resulting in microarray hybridization failure.Mol Biosyst. 2012 Apr;8(4):1325-38. doi: 10.1039/c2mb05300d. Epub 2012 Feb 7. Mol Biosyst. 2012. PMID: 22314967
-
Effects of DNA secondary structure on oligonucleotide probe binding efficiency.Comput Biol Chem. 2005 Dec;29(6):393-7. doi: 10.1016/j.compbiolchem.2005.09.002. Epub 2005 Nov 9. Comput Biol Chem. 2005. PMID: 16290040
-
Microarrays for identifying binding sites and probing structure of RNAs.Nucleic Acids Res. 2015 Jan;43(1):1-12. doi: 10.1093/nar/gku1303. Epub 2014 Dec 12. Nucleic Acids Res. 2015. PMID: 25505162 Free PMC article. Review.
-
Microarray oligonucleotide probes.Methods Enzymol. 2006;410:73-98. doi: 10.1016/S0076-6879(06)10004-X. Methods Enzymol. 2006. PMID: 16938547 Review.
Cited by
-
Properties of pseudo-complementary DNA substituted with weakly pairing analogs of guanine or cytosine.Nucleic Acids Res. 2008 Dec;36(22):6999-7008. doi: 10.1093/nar/gkn797. Epub 2008 Nov 5. Nucleic Acids Res. 2008. PMID: 18987000 Free PMC article.
-
A study of the relationships between oligonucleotide properties and hybridization signal intensities from NimbleGen microarray datasets.Nucleic Acids Res. 2008 May;36(9):2926-38. doi: 10.1093/nar/gkn133. Epub 2008 Apr 1. Nucleic Acids Res. 2008. PMID: 18385155 Free PMC article.
-
DNA hybridisation kinetics using single-molecule fluorescence imaging.Essays Biochem. 2021 Apr 16;65(1):27-36. doi: 10.1042/EBC20200040. Essays Biochem. 2021. PMID: 33491734 Free PMC article. Review.
-
Optimization and clinical validation of a pathogen detection microarray.Genome Biol. 2007;8(5):R93. doi: 10.1186/gb-2007-8-5-r93. Genome Biol. 2007. PMID: 17531104 Free PMC article.
-
A white-box approach to microarray probe response characterization: the BaFL pipeline.BMC Bioinformatics. 2009 Dec 29;10:449. doi: 10.1186/1471-2105-10-449. BMC Bioinformatics. 2009. PMID: 20040098 Free PMC article.
References
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. - DOI - PubMed
-
- Ramakrishnan R, Dorris D, Lublinsky A, Nguyen A, Domanus M, Prokhorova A, Gieser L, Touma E, Lockner R, Tata M, Zhu X, Patterson M, Shippy R, Sendera TJ, Mazumder A. An assessment of Motorola CodeLink microarray performance for gene expression profiling applications. Nucleic Acids Res. 2002;30:e30. doi: 10.1093/nar/30.7.e30. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

