Characterizing the function and structural organization of the 5' tRNA-like motif within the hepatitis C virus quasispecies
- PMID: 15755750
- PMCID: PMC1062876
- DOI: 10.1093/nar/gki290
Characterizing the function and structural organization of the 5' tRNA-like motif within the hepatitis C virus quasispecies
Abstract
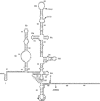
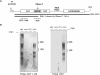
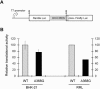
Hepatitis C virus (HCV) RNA is recognized and cleaved in vitro by RNase P enzyme near the AUG start codon. Because RNase P identifies transfer RNA (tRNA) precursors, it has been proposed that HCV RNA adopts structural similarities to tRNA. Here, we present experimental evidence of RNase P sensitivity conservation in natural RNA variant sequences, including a mutant sequence (A368-G) selected in vitro because it presented changes in the RNA structure of the relevant motif. The variation did not abrogate the original RNase P cleavage, but instead, it allowed a second cleavage at least 10 times more efficient, 4 nt downstream from the original one. The minimal RNA fragment that confers sensitivity to human RNase P enzyme was located between positions 299 and 408 (110 nt). Therefore, most of the tRNA-like domain resides within the viral internal ribosome entry site (IRES) element. In the variant, in which the mutation stabilizes a 4 nt stem-loop, the second cleavage required a shorter (60 nt) substrate, internal to the minimal fragment substrate, conforming a second tRNA-like structure with similarities to a 'Russian-doll' toy. This new structure did not impair IRES activity, albeit slightly reduced the efficiency of translation both in vitro and in transfected cells. Conservation of the original tRNA-like conformation together with preservation of IRES activity points to an essential role for this motif. This conservation is compatible with the presence of RNA structures with different complexity around the AUG start codon within a single viral population (quasispecies).
Figures







References
-
- Domingo E., Holland J.J. Mutation rates and rapid evolution of RNA viruses. In: Morse S.S., editor. Evolutionary Biology of Viruses. New York: Raven Press; 1994. pp. 161–184.
-
- Schuster P. Genotypes with phenotypes: adventures in an RNA toy world. Biophys. Chem. 1997;66:75–110. - PubMed
-
- Honda M., Ping L.H., Rijnbrand R.C., Amphlett E., Clarke B., Rowlands D., Lemon S.M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

