On the utility of pooling biological samples in microarray experiments
- PMID: 15755808
- PMCID: PMC552978
- DOI: 10.1073/pnas.0500607102
On the utility of pooling biological samples in microarray experiments
Abstract
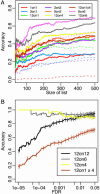
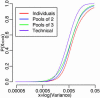
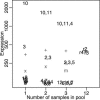
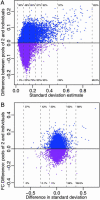
Over 15% of the data sets catalogued in the Gene Expression Omnibus Database involve RNA samples that have been pooled before hybridization. Pooling affects data quality and inference, but the exact effects are not yet known because pooling has not been systematically studied in the context of microarray experiments. Here we report on the results of an experiment designed to evaluate the utility of pooling and the impact on identifying differentially expressed genes. We find that inference for most genes is not adversely affected by pooling, and we recommend that pooling be done when fewer than three arrays are used in each condition. For larger designs, pooling does not significantly improve inferences if few subjects are pooled. The realized benefits in this case do not outweigh the price paid for loss of individual specific information. Pooling is beneficial when many subjects are pooled, provided that independent samples contribute to multiple pools.
Figures


References
-
- Jin, W., Riley, R. M., Wolfinger, R. D., White, K. P., Passador-Gurgel, G. & Gibson, G. (2001) Nat. Genet. 29, 389–395. - PubMed
-
- Saban, M. R., Hellmich, H., Nguyen, N., Winston, J., Hammond, T. G. & Saban, R. (2001) Physiol. Genomics 5, 147–160. - PubMed
-
- Chabas, D., Baranzini, S. E., Mitchell, D., Bernard, C. C., Rittling, S. R., Denhardt, D. T., Sobel, R. A., Lock, C., Karpuj, M., Pedotti, R., et al. (2001) Science 294, 1731–1735. - PubMed
-
- Waring, J. F., Jolly, R. A., Ciurlionis, R., Lum, P. Y., Praestgaard, J. T., Morfitt, D. C., Buratto, B., Roberts, C., Schadt, E. & Ulrich, R. G. (2001) Toxicol. Appl. Pharmacol. 175, 28–42. - PubMed
-
- Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. (2002) Science, 296, 340–343. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

