Functional evolution of a cis-regulatory module
- PMID: 15757364
- PMCID: PMC1064851
- DOI: 10.1371/journal.pbio.0030093
Functional evolution of a cis-regulatory module
Abstract
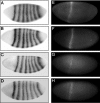
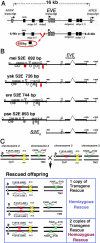
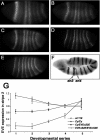
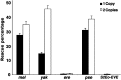
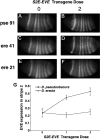
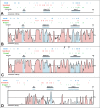
Lack of knowledge about how regulatory regions evolve in relation to their structure-function may limit the utility of comparative sequence analysis in deciphering cis-regulatory sequences. To address this we applied reverse genetics to carry out a functional genetic complementation analysis of a eukaryotic cis-regulatory module-the even-skipped stripe 2 enhancer-from four Drosophila species. The evolution of this enhancer is non-clock-like, with important functional differences between closely related species and functional convergence between distantly related species. Functional divergence is attributable to differences in activation levels rather than spatiotemporal control of gene expression. Our findings have implications for understanding enhancer structure-function, mechanisms of speciation and computational identification of regulatory modules.
Figures

References
-
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: Molecular genetics and the evolution of animal design. Oxford: Blackwell; 2001. 214 pp.
-
- Ludwig MZ. Functional evolution of noncoding DNA. Curr Opin Genet Dev. 2002;12:634–639. - PubMed
-
- Stone JR, Wray GA. Rapid evolution of cis-regulatory sequences via local point mutations. Mol Biol Evol. 2001;18:1764–1770. - PubMed
-
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

