Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion
- PMID: 15757514
- PMCID: PMC1084347
- DOI: 10.1186/1471-2180-5-9
Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion
Abstract
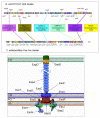
Background: Like many other pathogens, enterohaemorrhagic and enteropathogenic strains of Escherichia coli employ a type-III secretion system to translocate bacterial effector proteins into host cells, where they then disrupt a range of cellular functions. This system is encoded by the locus for enterocyte effacement. Many of the genes within this locus have been assigned names and functions through homology with the better characterised Ysc-Yop system from Yersinia spp. However, the functions and homologies of many LEE genes remain obscure.
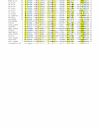
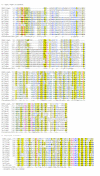
Results: We have performed a fresh bioinformatics analysis of the LEE. Using PSI-BLAST we have been able to identify several novel homologies between LEE-encoded and Ysc-Yop-associated proteins: Orf2/YscE, Orf5/YscL, rORF8/EscI, SepQ/YscQ, SepL/YopN-TyeA, CesD2/LcrR. In addition, we highlight homology between EspA and flagellin, and report many new homologues of the chaperone CesT.
Conclusion: We conclude that the vast majority of LEE-encoded proteins do indeed possess homologues and that homology data can be used in combination with experimental data to make fresh functional predictions.
Figures



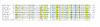


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

