Opinion: Cell entry machines: a common theme in nature?
- PMID: 15759040
- PMCID: PMC7097853
- DOI: 10.1038/nrmicro1131
Opinion: Cell entry machines: a common theme in nature?
Abstract
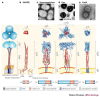
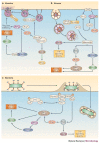
Molecular machines orchestrate the translocation and entry of pathogens through host cell membranes, in addition to the uptake and release of molecules during endocytosis and exocytosis. Viral cell entry requires a family of glycoproteins, and the structural organization and function of these viral glycoproteins are similar to the SNARE proteins, which are known to be involved in intracellular vesicle fusion, endocytosis and exocytosis. Here, we propose that a family of bacterial membrane proteins that are responsible for cell-mediated adherence and entry resembles the structural architecture of both viral fusion proteins and eukaryotic SNAREs and might therefore share similar, but distinct, mechanisms of cell membrane translocation. Furthermore, we propose that the recurrence of these molecular machines across species indicates that these architectural motifs were evolutionarily selected because they provided the best solution to ensure the survival of pathogens within a particular environment.
Conflict of interest statement
The authors are employed by Chiron Vaccines
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

