Identifying candidate Hirschsprung disease-associated RET variants
- PMID: 15759212
- PMCID: PMC1199373
- DOI: 10.1086/429589
Identifying candidate Hirschsprung disease-associated RET variants
Abstract
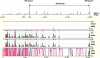
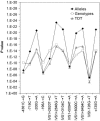
Patients with sporadic Hirschsprung disease (HSCR) show increased allele sharing at markers in the 5' region of the RET locus, indicating the presence of a common ancestral RET mutation. In a previous study, we found a haplotype of six SNPs that was transmitted to 55.6% of our patients, whereas it was present in only 16.2% of the controls we used. Among the patients with that haplotype, 90.8% had it on both chromosomes, which led to a much higher risk of developing HSCR than when the haplotype occurred heterozygously. To more precisely define the HSCR-associated region and to identify candidate disease-associated variant(s), we sequenced the shared common haplotype region from 10 kb upstream of the RET gene through intron 1 and exon 2 (in total, 33 kb) in a patient homozygous for the common risk haplotype and in a control individual homozygous for the most common nonrisk haplotype. A comparison of these sequences revealed 86 sequence differences. Of these 86 variations, 8 proved to be in regions highly conserved among different vertebrates and within putative transcription factor binding sites. We therefore considered these as candidate disease-associated variants. Subsequent genotyping of these eight variants revealed a strong disease association for six of the eight markers. These six markers also showed the largest distortions in allele transmission. Interspecies comparison showed that only one of the six variations was located in a region also conserved in a nonmammalian species, making it the most likely candidate HSCR-associated variant.
Figures
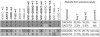
References
Electronic-Database Information
-
- Hirschsprung Disease, University of Groningen Department of Medical Genetics, http://www.rug.nl/med/faculteit/disciplinegroepen/medischegenetica/hered... (for additional information about HSCR)
-
- MULAN, http://mulan.dcode.org/
-
- NCBI Home Page, http://www.ncbi.nlm.nih.gov/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HSCR and RET)
-
- UCSC Genome Browser, http://genome.ucsc.edu/
References
-
- Bolk S, Pelet A, Hofstra RMW, Angrist M, Salomon R, Croaker D, Buys CHCM, Lyonnet S, Chakravarti A (2000) A human model for multigenic inheritance: phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci USA 97:268–27310.1073/pnas.97.1.268 - DOI - PMC - PubMed
-
- Borrego S, Wright F, Fernandez R, Williams N, Lopez-Alonso M, Davuluri R, Antinolo G, Eng C (2003) A founding locus within the RET proto-oncogene may account for a large proportion of apparently sporadic Hirschsprung disease and a subset of cases of sporadic medullary thyroid carcinoma. Am J Hum Genet 72:88–100 - PMC - PubMed
-
- Burzynski GM, Nolte IM, Osinga J, Ceccherini I, Twigt B, Maas S, Brooks A, Verheij J, Plaza Menacho I, Buys CHCM, Hofstra RMW (2004) Localizing a putative mutation as the major contributor to the development of sporadic Hirschsprung disease to the RET genomic sequence between the promoter region and exon 2. Eur J Hum Genet 12:604–61210.1038/sj.ejhg.5201199 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

