CoPub Mapper: mining MEDLINE based on search term co-publication
- PMID: 15760478
- PMCID: PMC1274248
- DOI: 10.1186/1471-2105-6-51
CoPub Mapper: mining MEDLINE based on search term co-publication
Abstract
Background: High throughput microarray analyses result in many differentially expressed genes that are potentially responsible for the biological process of interest. In order to identify biological similarities between genes, publications from MEDLINE were identified in which pairs of gene names and combinations of gene name with specific keywords were co-mentioned.
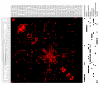
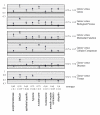
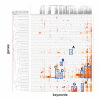
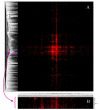
Results: MEDLINE search strings for 15,621 known genes and 3,731 keywords were generated and validated. PubMed IDs were retrieved from MEDLINE and relative probability of co-occurrences of all gene-gene and gene-keyword pairs determined. To assess gene clustering according to literature co-publication, 150 genes consisting of 8 sets with known connections (same pathway, same protein complex, or same cellular localization, etc.) were run through the program. Receiver operator characteristics (ROC) analyses showed that most gene sets were clustered much better than expected by random chance. To test grouping of genes from real microarray data, 221 differentially expressed genes from a microarray experiment were analyzed with CoPub Mapper, which resulted in several relevant clusters of genes with biological process and disease keywords. In addition, all genes versus keywords were hierarchical clustered to reveal a complete grouping of published genes based on co-occurrence.
Conclusion: The CoPub Mapper program allows for quick and versatile querying of co-published genes and keywords and can be successfully used to cluster predefined groups of genes and microarray data.
Figures


Similar articles
-
CoPub: a literature-based keyword enrichment tool for microarray data analysis.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W406-10. doi: 10.1093/nar/gkn215. Epub 2008 Apr 28. Nucleic Acids Res. 2008. PMID: 18442992 Free PMC article.
-
MILANO--custom annotation of microarray results using automatic literature searches.BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article.
-
PSE: a tool for browsing a large amount of MEDLINE/PubMed abstracts with gene names and common words as the keywords.BMC Bioinformatics. 2005 Dec 10;6:295. doi: 10.1186/1471-2105-6-295. BMC Bioinformatics. 2005. PMID: 16336692 Free PMC article.
-
An overview of Spotfire for gene-expression studies.Curr Protoc Bioinformatics. 2004 Sep;Chapter 7:Unit 7.7. doi: 10.1002/0471250953.bi0707s6. Curr Protoc Bioinformatics. 2004. PMID: 18428733 Review.
-
Matrix factorisation methods applied in microarray data analysis.Int J Data Min Bioinform. 2010;4(1):72-90. doi: 10.1504/ijdmb.2010.030968. Int J Data Min Bioinform. 2010. PMID: 20376923 Free PMC article. Review.
Cited by
-
Discovery of disease- and drug-specific pathways through community structures of a literature network.Bioinformatics. 2020 Mar 1;36(6):1881-1888. doi: 10.1093/bioinformatics/btz857. Bioinformatics. 2020. PMID: 31738408 Free PMC article.
-
Functional variants identify sex-specific genes and pathways in Alzheimer's Disease.Nat Commun. 2023 May 13;14(1):2765. doi: 10.1038/s41467-023-38374-z. Nat Commun. 2023. PMID: 37179358 Free PMC article.
-
Gene regulatory networks in lactation: identification of global principles using bioinformatics.BMC Syst Biol. 2007 Nov 27;1:56. doi: 10.1186/1752-0509-1-56. BMC Syst Biol. 2007. PMID: 18039394 Free PMC article.
-
CoPub: a literature-based keyword enrichment tool for microarray data analysis.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W406-10. doi: 10.1093/nar/gkn215. Epub 2008 Apr 28. Nucleic Acids Res. 2008. PMID: 18442992 Free PMC article.
-
Linking genes to literature: text mining, information extraction, and retrieval applications for biology.Genome Biol. 2008;9 Suppl 2(Suppl 2):S8. doi: 10.1186/gb-2008-9-s2-s8. Epub 2008 Sep 1. Genome Biol. 2008. PMID: 18834499 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

