Evolutionary sequence analysis of complete eukaryote genomes
- PMID: 15762985
- PMCID: PMC1274250
- DOI: 10.1186/1471-2105-6-53
Evolutionary sequence analysis of complete eukaryote genomes
Abstract
Background: Gene duplication and gene loss during the evolution of eukaryotes have hindered attempts to estimate phylogenies and divergence times of species. Although current methods that identify clusters of orthologous genes in complete genomes have helped to investigate gene function and gene content, they have not been optimized for evolutionary sequence analyses requiring strict orthology and complete gene matrices. Here we adopt a relatively simple and fast genome comparison approach designed to assemble orthologs for evolutionary analysis. Our approach identifies single-copy genes representing only species divergences (panorthologs) in order to minimize potential errors caused by gene duplication. We apply this approach to complete sets of proteins from published eukaryote genomes specifically for phylogeny and time estimation.
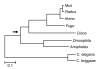
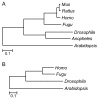
Results: Despite the conservative criterion used, 753 panorthologs (proteins) were identified for evolutionary analysis with four genomes, resulting in a single alignment of 287,000 amino acids. With this data set, we estimate that the divergence between deuterostomes and arthropods took place in the Precambrian, approximately 400 million years before the first appearance of animals in the fossil record. Additional analyses were performed with seven, 12, and 15 eukaryote genomes resulting in similar divergence time estimates and phylogenies.
Conclusion: Our results with available eukaryote genomes agree with previous results using conventional methods of sequence data assembly from genomes. They show that large sequence data sets can be generated relatively quickly and efficiently for evolutionary analyses of complete genomes.
Figures


References
-
- House CH, Runnegar B, Fitz-Gibbon ST. Geobiological analysis using whole genome-based tree building applied to the Bacteria, Archaea, and Eukarya. Geobiology. 2003;1:15–26. doi: 10.1046/j.1472-4669.2003.00004.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

