Gene expression signature of estrogen receptor alpha status in breast cancer
- PMID: 15762987
- PMCID: PMC555753
- DOI: 10.1186/1471-2164-6-37
Gene expression signature of estrogen receptor alpha status in breast cancer
Abstract
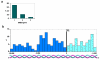
Background: Estrogens are known to regulate the proliferation of breast cancer cells and to modify their phenotypic properties. Identification of estrogen-regulated genes in human breast tumors is an essential step toward understanding the molecular mechanisms of estrogen action in cancer. To this end we generated and compared the Serial Analysis of Gene Expression (SAGE) profiles of 26 human breast carcinomas based on their estrogen receptor alpha (ER) status. Thus, producing a breast cancer SAGE database of almost 2.5 million tags, representing over 50,000 transcripts.
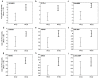
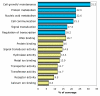
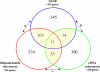
Results: We identified 520 transcripts differentially expressed between ERalpha-positive (+) and ERalpha-negative (-) primary breast tumors (Fold change >or= 2; p < 0.05). Furthermore, we identified 220 high-affinity Estrogen Responsive Elements (EREs) distributed on the promoter regions of 163 out of the 473 up-modulated genes in ERalpha (+) breast tumors. In brief, we observed predominantly up-regulation of cell growth related genes, DNA binding and transcription factor activity related genes based on Gene Ontology (GO) biological functional annotation. GO terms over-representation analysis showed a statistically significant enrichment of various transcript families including: metal ion binding related transcripts (p = 0.011), calcium ion binding related transcripts (p = 0.033) and steroid hormone receptor activity related transcripts (p = 0.031). SAGE data associated with ERalpha status was compared with reported information from breast cancer DNA microarrays studies. A significant proportion of ERalpha associated gene expression changes was validated by this cross-platform comparison. However, our SAGE study also identified novel sets of genes as highly expressed in ERalpha (+) invasive breast tumors not previously reported. These observations were further validated in an independent set of human breast tumors by means of real time RT-PCR.
Conclusion: The integration of the breast cancer comparative transcriptome analysis based on ERalpha status coupled to the genome-wide identification of high-affinity EREs and GO over-representation analysis, provide useful information for validation and discovery of signaling networks related to estrogen response in this malignancy.
Figures
References
-
- Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;296:1642–1644. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

