Determination of barrier heights and prefactors from protein folding rate data
- PMID: 15764665
- PMCID: PMC1305610
- DOI: 10.1529/biophysj.104.052548
Determination of barrier heights and prefactors from protein folding rate data
Abstract
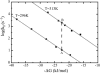
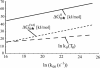
We determine both barrier heights and prefactors for protein folding by applying constraints determined from experimental rate measurements to a Kramers theory for folding rate. The theoretical values are required to match the experimental values at two conditions of temperature and denaturant that induce the same stability. Several expressions for the prefactor in the Kramers rate equation are examined: a random energy approximation, a correlated energy approximation, and an approximation using a single Arrhenius activation energy. Barriers and prefactors are generally found to be large as a result of implementing this recipe, i.e., the folding landscape is cooperative and smooth. Interestingly, a prefactor with a single Arrhenius activation energy admits no formal solution.
Figures



References
-
- Akmal, A., and V. Munoz. 2004. The nature of the free energy barriers to two state folding. Proteins. 57:142–152. - PubMed
-
- Bryngelson, J. D., J. N. Onuchic, N. D. Socci, and P. G. Wolynes. 1995. Funnels, pathways and the energy landscape of protein folding. Proteins. 21:167–195. - PubMed
-
- Bryngelson, J. D., and P. G. Wolynes. 1989. Intermediates and barrier crossing in a random energy model (with applications to protein folding). J. Phys. Chem. 93:6902–6915.
-
- CRC. 2003. CRC Handbook of Chemistry and Physics, 84th Ed. D.R. Lide, editor. CRC Press, New York.
-
- D'Aquino, J. A., J. Gomez, V. J. Hilser, K. H. Lee, L. M. Amzel, and E. Freire. 1996. The magnitude of the backbone conformational entropy change in protein folding. Proteins Struct. Funct. Gen. 25:143–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

