Genetic causes of human heart failure
- PMID: 15765133
- PMCID: PMC1052010
- DOI: 10.1172/JCI24351
Genetic causes of human heart failure
Abstract
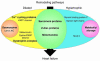
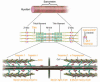
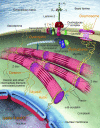
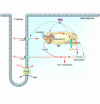
Factors that render patients with cardiovascular disease at high risk for heart failure remain incompletely defined. Recent insights into molecular genetic causes of myocardial diseases have highlighted the importance of single-gene defects in the pathogenesis of heart failure. Through analyses of the mechanisms by which a mutation selectively perturbs one component of cardiac physiology and triggers cell and molecular responses, studies of human gene mutations provide a window into the complex processes of cardiac remodeling and heart failure. Knowledge gleaned from these studies shows promise for defining novel therapeutic targets for genetic and acquired causes of heart failure.
Figures

References
-
- American Heart Association. 2004. Heart disease and stroke statistics – 2004 update. American Heart Association. http://www.americanheart.org.
-
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. - PubMed
-
- CardioGenomics. Genomics of cardiovascular development, adaptation, and remodeling. NHLBI program for genomic applications. Harvard Medical School. http://cardiogenomics.med.harvard.edu.
-
- Morita H, et al. Molecular epidemiology of hypertrophic cardiomyopathy. Cold Spring Harb. Symp. Quant. Biol. 2002;67:383–388. - PubMed
-
- Richard P, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

