Speeding disease gene discovery by sequence based candidate prioritization
- PMID: 15766383
- PMCID: PMC1274252
- DOI: 10.1186/1471-2105-6-55
Speeding disease gene discovery by sequence based candidate prioritization
Abstract
Background: Regions of interest identified through genetic linkage studies regularly exceed 30 centimorgans in size and can contain hundreds of genes. Traditionally this number is reduced by matching functional annotation to knowledge of the disease or phenotype in question. However, here we show that disease genes share patterns of sequence-based features that can provide a good basis for automatic prioritization of candidates by machine learning.
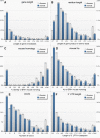
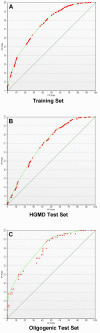
Results: We examined a variety of sequence-based features and found that for many of them there are significant differences between the sets of genes known to be involved in human hereditary disease and those not known to be involved in disease. We have created an automatic classifier called PROSPECTR based on those features using the alternating decision tree algorithm which ranks genes in the order of likelihood of involvement in disease. On average, PROSPECTR enriches lists for disease genes two-fold 77% of the time, five-fold 37% of the time and twenty-fold 11% of the time.
Conclusion: PROSPECTR is a simple and effective way to identify genes involved in Mendelian and oligogenic disorders. It performs markedly better than the single existing sequence-based classifier on novel data. PROSPECTR could save investigators looking at large regions of interest time and effort by prioritizing positional candidate genes for mutation detection and case-control association studies.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

