Recognition and binding of mismatch repair proteins at an oncogenic hot spot
- PMID: 15766387
- PMCID: PMC555755
- DOI: 10.1186/1471-2199-6-6
Recognition and binding of mismatch repair proteins at an oncogenic hot spot
Abstract
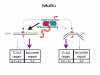
Background: The current investigation was undertaken to determine key steps differentiating G:T and G:A repair at the H-ras oncogenic hot spot within the nuclear environment because of the large difference in repair efficiency of these two mismatches.
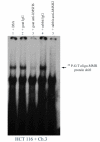
Results: Electrophoretic mobility shift (gel shift) experiments demonstrate that DNA containing mismatched bases are recognized and bound equally efficiently by hMutSalpha in both MMR proficient and MMR deficient (hMLH1-/-) nuclear extracts. Competition experiments demonstrate that while hMutSalpha predictably binds the G:T mismatch to a much greater extent than G:A, hMutSalpha demonstrates a surprisingly equal ratio of competitive inhibition for both G:T and G:A mismatch binding reactions at the H-ras hot spot of mutation. Further, mismatch repair assays reveal almost 2-fold higher efficiency of overall G:A repair (5'-nick directed correct MMR to G:C and incorrect repair to T:A), as compared to G:T overall repair. Conversely, correct MMR of G:T --> G:C is significantly higher (96%) than that of G:A --> G:C (60%).
Conclusion: Combined, these results suggest that initiation of correct MMR requires the contribution of two separate steps; initial recognition by hMutSalpha followed by subsequent binding. The 'avidity' of the binding step determines the extent of MMR pathway activation, or the activation of a different cellular pathway. Thus, initial recognition by hMutSalpha in combination with subsequent decreased binding to the G:A mismatch (as compared to G:T) may contribute to the observed increased frequency of incorrect repair of G:A, resulting in the predominant GGC --> GTC (Gly --> Val) ras-activating mutation found in a high percentage of human tumors.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

