Identification of the 'NORE' (N-Oct-3 responsive element), a novel structural motif and composite element
- PMID: 15767276
- PMCID: PMC1065252
- DOI: 10.1093/nar/gki284
Identification of the 'NORE' (N-Oct-3 responsive element), a novel structural motif and composite element
Abstract
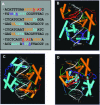
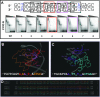
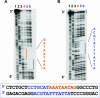
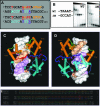
N-Oct-3 is a neuronal transcription factor widely expressed in the developing mammalian central nervous system, and necessary to maintain neural cell differentiation. The key role of N-Oct-3 in the transcriptional regulation of a multiplicity of genes is primarily due to the structural plasticity of its so-called 'POU' (acronym of Pit, Oct, Unc) DNA-binding domain. We have recently reported about the unusual dual neuro-specific transcriptional regulation displayed by N-Oct-3 [Blaud,M., Vossen,C., Joseph,G., Alazard,R., Erard,M. and Nieto,L. (2004) J. Mol. Biol., 339, 1049-1058]. To elucidate the underlying molecular mechanisms, we have now made use of molecular modeling, DNA footprinting and electrophoretic mobility shift assay techniques. This combined approach has allowed us to uncover a novel mode of homodimerization adopted by the N-Oct-3 POU domain bound to the neuronal aromatic amino acids de-carboxylase and corticotropin-releasing hormone gene promoters and to demonstrate that this pattern is induced by a structural motif that we have termed 'NORE' (N-Oct-3 responsive element), comprising the 14 bp sequence element TNNRTAAATAATRN. In addition, we have been able to explain how the same structural motif can also induce the formation of a heterodimer in association with hepatocyte nuclear factor 3beta(/Forkhead box a2). Finally, we discuss the possible role of the NORE motif in relation to neuroendocrine lung tumor formation, and in particular the development of small cell lung cancer.
Figures
References
-
- Warren A.J. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 2002;12:107–114. - PubMed
-
- Johnson W., Jameson J.L. Transcriptional control of gene expression. In: Jameson J.L., editor. Principles of Molecular Medicine. Towota, NJ: Humana Press Inc.; 1998.
-
- Latchman D.S. Transcription factors as potential targets for therapeutic drugs. Curr. Pharm. Biotechnol. 2000;1:57–61. - PubMed

