Dimeric configuration of SeqA protein bound to a pair of hemi-methylated GATC sequences
- PMID: 15767277
- PMCID: PMC1065253
- DOI: 10.1093/nar/gki289
Dimeric configuration of SeqA protein bound to a pair of hemi-methylated GATC sequences
Abstract
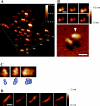
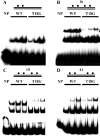
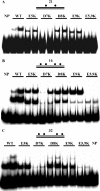
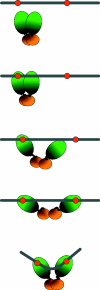
The binding of SeqA protein to hemi-methylated GATC sequences (hemi-sites) regulates chromosome initiation and the segregation of replicated chromosome in Escherichia coli. We have used atomic force microscopy to examine the architecture of SeqA and the mode of binding of one molecule of SeqA to a pair of hemi-sites in aqueous solution. SeqA has a bipartite structure composed of a large and a small lobe. Upon binding of a SeqA molecule to a pair of hemi-sites, the larger lobe becomes visibly separated into two DNA binding domains, each of which binds to one hemi-site. The two DNA binding domains are held together by association between the two multimerization domains that make up the smaller lobe. The binding of each DNA binding domain to a hemi-site leads to bending of the bound DNA inwards toward the bound protein. In this way, SeqA adopts a dimeric configuration when bound to a pair of hemi-sites. Mutational analysis of the multimerization domain indicates that, in addition to multimerization of SeqA polypeptides, this domain contributes to the ability of SeqA to bind to a pair of hemi-sites and to its cooperative behavior.
Figures

Similar articles
-
Aggregation of SeqA protein requires positively charged amino acids in the hinge region.Biochem Biophys Res Commun. 2007 Aug 17;360(1):63-9. doi: 10.1016/j.bbrc.2007.05.225. Epub 2007 Jun 18. Biochem Biophys Res Commun. 2007. PMID: 17586464
-
Modeling of the full-length Escherichia coli SeqA protein, in complex with DNA.Pathol Biol (Paris). 2009 May;57(3):e61-6. doi: 10.1016/j.patbio.2008.03.013. Epub 2008 Oct 11. Pathol Biol (Paris). 2009. PMID: 18849124
-
Specific N-terminal interactions of the Escherichia coli SeqA protein are required to form multimers that restrain negative supercoils and form foci.Genes Cells. 2005 Nov;10(11):1039-49. doi: 10.1111/j.1365-2443.2005.00898.x. Genes Cells. 2005. PMID: 16236133
-
The Escherichia coli SeqA protein.Plasmid. 2009 May;61(3):141-50. doi: 10.1016/j.plasmid.2009.02.004. Epub 2009 Feb 28. Plasmid. 2009. PMID: 19254745 Review.
-
Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli.Biometals. 2007 Jun;20(3-4):263-74. doi: 10.1007/s10534-006-9060-9. Epub 2006 Dec 22. Biometals. 2007. PMID: 17186377 Review.
Cited by
-
DNA Methylation.EcoSal Plus. 2014 May;6(1):10.1128/ecosalplus.ESP-0003-2013. doi: 10.1128/ecosalplus.ESP-0003-2013. EcoSal Plus. 2014. PMID: 26442938 Free PMC article.
-
Mutant DnaAs of Escherichia coli that are refractory to negative control.Nucleic Acids Res. 2013 Dec;41(22):10254-67. doi: 10.1093/nar/gkt774. Epub 2013 Aug 29. Nucleic Acids Res. 2013. PMID: 23990329 Free PMC article.
-
Regulating DNA replication in bacteria.Cold Spring Harb Perspect Biol. 2013 Apr 1;5(4):a012922. doi: 10.1101/cshperspect.a012922. Cold Spring Harb Perspect Biol. 2013. PMID: 23471435 Free PMC article. Review.
-
The synchrony phenotype persists after elimination of multiple GATC sites from the dnaA promoter of Escherichia coli.J Bacteriol. 2006 Jun;188(12):4573-6. doi: 10.1128/JB.00089-06. J Bacteriol. 2006. PMID: 16740964 Free PMC article.
-
SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC.Mol Cell. 2006 Nov 17;24(4):581-92. doi: 10.1016/j.molcel.2006.09.016. Mol Cell. 2006. PMID: 17114060 Free PMC article.
References
-
- Sarraf S.A., Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15:595–605. - PubMed
-
- Wade P.A. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–1137. - PubMed
-
- Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. - PubMed
-
- Geier G.E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J. Biol. Chem. 1979;254:1408–1413. - PubMed
-
- Campbell J.L., Kleckner N. E.coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

