Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death
- PMID: 15767425
- PMCID: PMC1061538
- DOI: 10.1128/JVI.79.7.4238-4245.2005
Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death
Abstract
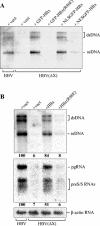
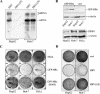
The hepatitis B virus (HBV) X protein (HBx) is essential for virus infection and has been implicated in the development of liver cancer associated with chronic infection. HBx can interact with a number of cellular proteins, and in cell culture, it exhibits pleiotropic activities, among which is its ability to interfere with cell viability and stimulate HBV replication. Previous work has demonstrated that HBx affects cell viability by a mechanism that requires its binding to DDB1, a highly conserved protein implicated in DNA repair and cell cycle regulation. We now show that an interaction with DDB1 is also needed for HBx to stimulate HBV genome replication. Thus, HBx point mutants defective for DDB1 binding fail to complement the low level of replication of an HBx-deficient HBV genome when provided in trans, and one such mutant regains activity when directly fused to DDB1. Furthermore, DDB1 depletion by RNA interference specifically compromises replication of wild-type HBV, indicating that HBx produced from the viral genome also functions in a DDB1-dependent fashion. We also show that HBx in association with DDB1 acts in the nucleus and stimulates HBV replication mainly by enhancing viral mRNA levels, regardless of whether the protein is expressed from the HBV genome itself or supplied in trans. Interestingly, whereas HBx induces cell death in both HepG2 and Huh-7 hepatoma cell lines, it enhances HBV replication only in HepG2 cells, suggesting that the two activities involve distinct DDB1-dependent pathways.
Figures


References
-
- Arbuthnot, P., A. Capovilla, and M. Kew. 2000. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J. Gastroenterol. Hepatol. 15:357-368. - PubMed
-
- Bondar, T., A. Ponomarev, and P. Raychaudhuri. 2004. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J. Biol. Chem. 279:9937-9943. - PubMed
-
- Bontron, S., N. Lin-Marq, and M. Strubin. 2002. Hepatitis B virus X protein associated with UV-DDB1 induces cell death in the nucleus and is functionally antagonized by UV-DDB2. J. Biol. Chem. 277:38847-38854. - PubMed
-
- Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

