Suppression of viral infectivity through lethal defection
- PMID: 15767582
- PMCID: PMC555496
- DOI: 10.1073/pnas.0408871102
Suppression of viral infectivity through lethal defection
Abstract
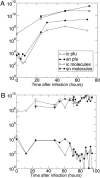
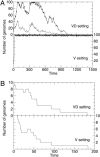
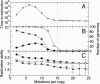
RNA viruses replicate with a very high error rate and give rise to heterogeneous, highly plastic populations able to adapt very rapidly to changing environments. Viral diseases are thus difficult to control because of the appearance of drug-resistant mutants, and it becomes essential to seek mechanisms able to force the extinction of the quasispecies before adaptation emerges. An alternative to the use of conventional drugs consists in increasing the replication error rate through the use of mutagens. Here, we report about persistent infections of lymphocytic choriomeningitis virus treated with fluorouracil, where a progressive debilitation of infectivity leading to eventual extinction occurs. The transition to extinction is accompanied by the production of large amounts of RNA, indicating that the replicative ability of the quasispecies is not strongly impaired by the mutagen. By means of experimental and theoretical approaches, we propose that a fraction of the RNA molecules synthesized can behave as a defective subpopulation able to drive the viable class extinct. Our results lead to the identification of two extinction pathways, one at high amounts of mutagen, where the quasispecies completely loses its ability to infect and replicate, and a second one, at lower amounts of mutagen, where replication continues while the infective class gets extinct because of the action of defectors. The results bear on a potential application of increased mutagenesis as an antiviral strategy in that low doses of a mutagenic agent may suffice to drive persistent virus to extinction.
Figures

References
-
- Domingo, E., Holland, J. J. (1988) in RNA Genetics, eds. Domingo, E., Holland, J. J. & Ahlquist, P. (CRC, Boca Raton, FL), Vol. 3, pp. 3–36.
-
- Biebricher, C. K. (1999) in Origin and Evolution of Viruses, eds. Domingo, E., Webster, R. K. & Holland, J. J. (Academic, San Diego), pp. 65–85.
-
- Eigen, M. & Schuster, P. (1977) Naturwissenschaften 64, 541–565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

