Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones
- PMID: 15767685
- PMCID: PMC1061639
- DOI: 10.1128/MCB.25.7.2819-2831.2005
Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones
Abstract
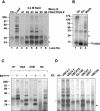
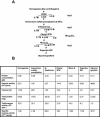
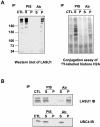
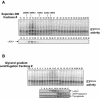
During spermatogenesis, a large fraction of cellular proteins is degraded as the spermatids evolve to their elongated mature forms. In particular, histones must be degraded in early elongating spermatids to permit chromatin condensation. Our laboratory previously demonstrated the activation of ubiquitin conjugation during spermatogenesis. This activation is dependent on the ubiquitin-conjugating enzyme (E2) UBC4, and a testis-particular isoform, UBC4-testis, is induced when histones are degraded. Therefore, we tested whether there are UBC4-dependent ubiquitin protein ligases (E3s) that can ubiquitinate histones. Indeed, a novel enzyme, E3Histone, which could conjugate ubiquitin to histones H1, H2A, H2B, H3, and H4 in vitro, was found. Only the UBC4/UBC5 family of E2s supported E3Histone-dependent ubiquitination of histone H2A, and of this family, UBC4-1 and UBC4-testis are the preferred E2s. We purified this ligase activity 3,600-fold to near homogeneity. Mass spectrometry of the final material revealed the presence of a 482-kDa HECT domain-containing protein, which was previously named LASU1. Anti-LASU1 antibodies immunodepleted E3Histone activity. Mass spectrometry and size analysis by gel filtration and glycerol gradient centrifugation suggested that E3Histone is a monomer of LASU1. Our assays also show that this enzyme is the major UBC4-1-dependent histone-ubiquitinating E3. E3Histone is therefore a HECT domain E3 that likely plays an important role in the chromatin condensation that occurs during spermatid maturation.
Figures




References
-
- Agell, N., M. Chiva, and C. Mezquita. 1983. Changes in nuclear content of protein conjugate histone H2A-ubiquitin during rooster spermatogenesis. FEBS Lett. 155:209-212. - PubMed
-
- Aravind, L. 2001. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26:273-275. - PubMed
-
- Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. - PubMed
-
- Beitel, L. K., Y. A. Elhaji, R. Lumbroso, S. S. Wing, V. Panet-Raymond, B. Gottlieb, L. Pinsky, and M. A. Trifiro. 2002. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J. Mol. Endocrinol. 29:41-60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
