Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export
- PMID: 15769879
- PMCID: PMC1370741
- DOI: 10.1261/rna.7291205
Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export
Abstract
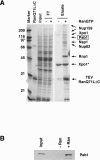
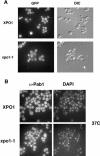
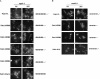
Pab1 is the major poly(A)-binding protein in yeast. It is a multifunctional protein that mediates many cellular functions associated with the 3'-poly(A)-tail of messenger RNAs. Here, we characterize Pab1 as an export cargo of the protein export factor Xpo1/Crm1. Pab1 is a major Xpo1/Crm1-interacting protein in yeast extracts and binds directly to Xpo1/Crm1 in a RanGTP-dependent manner. Pab1 shuttles rapidly between the nucleus and the cytoplasm and partially accumulates in the nucleus when the function of Xpo1/Crm1 is inhibited. However, Pab1 can also be exported by an alternative pathway, which is dependent on the MEX67-mRNA export pathway. Import of Pab1 is mediated by the import receptor Kap108/Sxm1 through a nuclear localization signal in its fourth RNA-binding domain. Interestingly, inhibition of Pab1's nuclear import causes a kinetic delay in the export of mRNA. Furthermore, the inviability of a pab1 deletion strain is suppressed by a mutation in the 5'-3' exoribonuclease RRP6, a component of the nuclear exosome. Therefore, nuclear Pab1 may be required for efficient mRNA export and may function in the quality control of mRNA in the nucleus.
Figures




References
-
- Afonina, E., Stauber, R., and Pavlakis, G.N. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273: 13015–13021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
