Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts
- PMID: 15772282
- PMCID: PMC1087999
- DOI: 10.1105/tpc.104.030452
Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts
Abstract
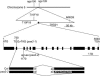
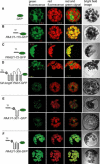
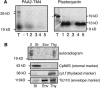
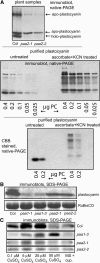
Copper delivery to the thylakoid lumen protein plastocyanin and the stromal enzyme Cu/Zn superoxide dismutase in chloroplasts is required for photosynthesis and oxidative stress protection. The copper delivery system in chloroplasts was characterized by analyzing the function of copper transporter genes in Arabidopsis thaliana. Two mutant alleles were identified of a previously uncharacterized gene, PAA2 (for P-type ATPase of Arabidopsis), which is required for efficient photosynthetic electron transport. PAA2 encodes a copper-transporting P-type ATPase with sequence similarity to PAA1, which functions in copper transport in chloroplasts. Both proteins localized to the chloroplast, as indicated by fusions to green fluorescent protein. The PAA1 fusions were found in the chloroplast periphery, whereas PAA2 fusions were localized in thylakoid membranes. The phenotypes of paa1 and paa2 mutants indicated that the two transporters have distinct functions: whereas both transporters are required for copper delivery to plastocyanin, copper delivery to the stroma is inhibited only in paa1 but not in paa2. The effects of paa1 and paa2 on superoxide dismutase isoform expression levels suggest that stromal copper levels regulate expression of the nuclear genes IRON SUPEROXIDE DISMUTASE1 and COPPER/ZINC SUPEROXIDE DISMUTASE2. A paa1 paa2 double mutant was seedling-lethal, underscoring the importance of copper to photosynthesis. We propose that PAA1 and PAA2 function sequentially in copper transport over the envelope and thylakoid membrane, respectively.
Figures







References
-
- Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygen and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. - PubMed
-
- Askwith, C., Eide, D., Van Ho, A., Bernard, P.S., Li, L., Davis-Kaplan, S., Sipe, D.M., and Kaplan, J. (1994). The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76, 403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

