Familial Alzheimer's disease presenilin 1 mutations cause alterations in the conformation of presenilin and interactions with amyloid precursor protein
- PMID: 15772361
- PMCID: PMC6725136
- DOI: 10.1523/JNEUROSCI.0364-05.2005
Familial Alzheimer's disease presenilin 1 mutations cause alterations in the conformation of presenilin and interactions with amyloid precursor protein
Abstract
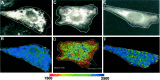
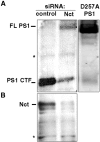
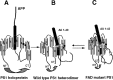
Presenilin 1 (PS1) is a critical component of the gamma-secretase complex, an enzymatic activity that cleaves amyloid beta (Abeta) from the amyloid precursor protein (APP). More than 100 mutations spread throughout the PS1 molecule are linked to autosomal dominant familial Alzheimer's disease (FAD). All of these mutations lead to a similar phenotype: an increased ratio of Abeta42 to Abeta40, increased plaque deposition, and early age of onset. We use a recently developed microscopy approach, fluorescence lifetime imaging microscopy, to monitor the relative molecular distance between PS1 N and C termini in intact cells. We show that FAD-linked missense mutations located near the N and C termini, in the mid-region of PS1, and the exon 9 deletion mutation all change the spatial relationship between PS1 N and C termini in a similar way, increasing proximity of the two epitopes. This effect is opposite of that observed by treatment with Abeta42-lowering nonsteroidal anti-inflammatory drugs (NSAIDs) (Lleo et al., 2004b). Accordingly, treatment of M146L PS1-overexpressing neurons with high-dose NSAIDs somewhat offsets the conformational change associated with the mutation. Moreover, by monitoring the relative distance between a PS1 loop epitope and the APP C terminus, we demonstrate that the FAD PS1 mutations are also associated with a consistent change in the configuration of the PS1-APP complex. The nonpathogenic E318G PS1 polymorphism had no effect on PS1 N terminus-C terminus proximity or PS1-APP interactions. We propose that the conformational change we observed may therefore provide a shared molecular mechanism for FAD pathogenesis caused by a wide range of PS1 mutations.
Figures


References
-
- Aldudo J, Bullido MJ, Frank A, Valdivieso F (1998) Missense mutation E318G of the presenilin-1 gene appears to be a nonpathogenic polymorphism. Ann Neurol 44: 985-986. - PubMed
-
- Annaert WG, Esselens C, Baert V, Boeve C, Snellings G, Cupers P, Craessaerts K, De Strooper B (2001) Interaction with telencephalin and the amyloid precursor protein predicts a ring structure for presenilins. Neuron 32: 579-589. - PubMed
-
- Bacskai BJ, Skoch J, Hickey GA, Allen R, Hyman BT (2003) FRET determinations using multiphoton fluorescence lifetime imaging microscopy (FLIM) to characterize amyloid-beta plaques. J Biomed Opt 8: 368-375. - PubMed
-
- Berezovska O, Frosch M, McLean P, Knowles R, Koo E, Kang D, Shen J, Lu FM, Lux SE, Tonegawa S, Hyman BT (1999) The Alzheimer-related gene presenilin 1 facilitates notch 1 in primary mammalian neurons. Brain Res Mol Brain Res 69: 273-280. - PubMed
-
- Berezovska O, Jack C, McLean P, Aster JC, Hicks C, Xia W, Wolfe MS, Kimberly WT, Weinmaster G, Selkoe DJ, Hyman BT (2000) Aspartate mutations in presenilin and gamma-secretase inhibitors both impair notch1 proteolysis and nuclear translocation with relative preservation of notch1 signaling. J Neurochem 75: 583-593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
