siRNA targeting the leader sequence of SARS-CoV inhibits virus replication
- PMID: 15772689
- PMCID: PMC7091583
- DOI: 10.1038/sj.gt.3302479
siRNA targeting the leader sequence of SARS-CoV inhibits virus replication
Abstract
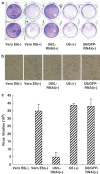
SARS-CoV (the SARS-Associated Coronavirus) was reported as a novel virus member in the coronavirus family, which was the cause of severe acute respiratory syndrome. Coronavirus replication occurs through a unique mechanism employing Leader sequence in the transcripts when initiating transcription from the genome. Therefore, we cloned the Leader sequence from SARS-CoV(BJ01), which is identical to that identified from SARS-CoV(HKU-39849), and constructed specific siRNA targeting the Leader sequence. Using EGFP and RFP reporter genes fused with the cloned SARS-CoV Leader sequence, we demonstrated that the siRNA targeting the Leader sequence decreased the mRNA abundance and protein expression levels of the reporter genes in 293T cells. By stably expressing the siRNA in Vero E6 cells, we provided data that the siRNA could effectively and specifically decrease the mRNA abundance of SARS-CoV genes as analyzed by RT-PCR and Northern blot. Our data indicated that the siRNA targeting the Leader sequence inhibited the replication of SARS-CoV in Vero E6 cells by silencing gene expression. We further demonstrated, via transient transfection experiments, that the siRNA targeting the Leader sequence had a much stronger inhibitory effect on SARS-CoV replication than the siRNAs targeting the Spike gene or the antisense oligodeoxynucleotides did. This report provides evidence that targeting Leader sequence using siRNA could be a powerful tool in inhibiting SARS-CoV replication.
Figures








Similar articles
-
Inhibition of SARS-CoV-2 Replication by a Small Interfering RNA Targeting the Leader Sequence.Viruses. 2021 Oct 8;13(10):2030. doi: 10.3390/v13102030. Viruses. 2021. PMID: 34696460 Free PMC article.
-
Inhibition of replication and infection of severe acute respiratory syndrome-associated coronavirus with plasmid-mediated interference RNA.Antivir Ther. 2005;10(4):527-33. Antivir Ther. 2005. PMID: 16038478
-
Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S.Antiviral Res. 2007 Mar;73(3):219-27. doi: 10.1016/j.antiviral.2006.10.008. Epub 2006 Nov 7. Antiviral Res. 2007. PMID: 17112601 Free PMC article.
-
Targeting the HIV-1 RNA leader sequence with synthetic oligonucleotides and siRNA: chemistry and cell delivery.Biochim Biophys Acta. 2006 Mar;1758(3):290-300. doi: 10.1016/j.bbamem.2005.10.013. Epub 2005 Nov 18. Biochim Biophys Acta. 2006. PMID: 16337923 Review.
-
[The research and application of inhibitory effect of RNA-based strategies on SARS].Bing Du Xue Bao. 2010 Jul;26(4):336-9. Bing Du Xue Bao. 2010. PMID: 20836389 Review. Chinese. No abstract available.
Cited by
-
Strategies for Targeting SARS CoV-2: Small Molecule Inhibitors-The Current Status.Front Immunol. 2020 Sep 18;11:552925. doi: 10.3389/fimmu.2020.552925. eCollection 2020. Front Immunol. 2020. PMID: 33072093 Free PMC article. Review.
-
SARS-CoV-2 Spike Protein Binding of Glycated Serum Albumin-Its Potential Role in the Pathogenesis of the COVID-19 Clinical Syndromes and Bias towards Individuals with Pre-Diabetes/Type 2 Diabetes and Metabolic Diseases.Int J Mol Sci. 2022 Apr 8;23(8):4126. doi: 10.3390/ijms23084126. Int J Mol Sci. 2022. PMID: 35456942 Free PMC article.
-
Inhibition of SARS-CoV-2 Replication by a Small Interfering RNA Targeting the Leader Sequence.Viruses. 2021 Oct 8;13(10):2030. doi: 10.3390/v13102030. Viruses. 2021. PMID: 34696460 Free PMC article.
-
COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment.Postgrad Med J. 2021 May;97(1147):312-320. doi: 10.1136/postgradmedj-2020-138577. Epub 2020 Sep 25. Postgrad Med J. 2021. PMID: 32978337 Free PMC article. Review.
-
Targeting herpetic keratitis by gene therapy.J Ophthalmol. 2012;2012:594869. doi: 10.1155/2012/594869. Epub 2012 Dec 26. J Ophthalmol. 2012. PMID: 23326647 Free PMC article.
References
-
- Blackmore T. Severe acute respiratory syndrome: a storm in a teacup? N Z Med J. 2003;116:U397. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

