Sequence changes in predicted promoter elements of STK11/LKB1 are unlikely to contribute to Peutz-Jeghers syndrome
- PMID: 15774015
- PMCID: PMC1084245
- DOI: 10.1186/1471-2164-6-38
Sequence changes in predicted promoter elements of STK11/LKB1 are unlikely to contribute to Peutz-Jeghers syndrome
Abstract
Background: Germline mutations or large-scale deletions in the coding region and splice sites of STK11/LKB1 do not account for all cases of Peutz-Jeghers syndrome (PJS). It is conceivable that, on the basis of data from other diseases, inherited variation in promoter elements of STK11/LKB1 may cause PJS.
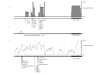
Results: Phylogenetic foot printing and transcription factor binding site prediction of sequence 5' to the coding sequence of STK11/LKB1 was performed to identify non-coding sequences of DNA indicative of regulatory elements. A series of 33 PJS cases in whom no mutation in STK11/LKB1 could be identified were screened for sequence changes in the putative promoter defined by nucleotides -1090 to -1472. Two novel sequence changes were identified, but were found to be present in healthy individuals.
Conclusion: These findings indicate that promoter sequence changes are unlikely to contribute to PJS.
Figures
References
-
- Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la CA, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

