Protein folding: the stepwise assembly of foldon units
- PMID: 15774579
- PMCID: PMC555724
- DOI: 10.1073/pnas.0501043102
Protein folding: the stepwise assembly of foldon units
Abstract
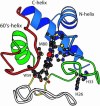
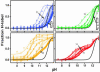
Equilibrium and kinetic hydrogen exchange experiments show that cytochrome c is composed of five foldon units that continually unfold and refold even under native conditions. Folding proceeds by the stepwise assembly of the foldon units rather than one amino acid at a time. The folding pathway is determined by a sequential stabilization process; previously formed foldons guide and stabilize subsequent foldons to progressively build the native protein. Four other proteins have been found to show similar behavior. These results support stepwise protein folding pathways through discrete intermediates.
Figures

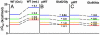



Similar articles
-
Branching in the sequential folding pathway of cytochrome c.Protein Sci. 2007 Sep;16(9):1946-56. doi: 10.1110/ps.072922307. Epub 2007 Jul 27. Protein Sci. 2007. PMID: 17660254 Free PMC article.
-
Order of steps in the cytochrome C folding pathway: evidence for a sequential stabilization mechanism.J Mol Biol. 2006 Jun 23;359(5):1410-9. doi: 10.1016/j.jmb.2006.04.035. Epub 2006 May 2. J Mol Biol. 2006. PMID: 16690080
-
Functional role of a protein foldon--an Omega-loop foldon controls the alkaline transition in ferricytochrome c.Proteins. 2006 May 1;63(2):349-55. doi: 10.1002/prot.20757. Proteins. 2006. PMID: 16287119
-
The nature of protein folding pathways.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):15873-80. doi: 10.1073/pnas.1411798111. Epub 2014 Oct 17. Proc Natl Acad Sci U S A. 2014. PMID: 25326421 Free PMC article. Review.
-
Protein folding and misfolding: mechanism and principles.Q Rev Biophys. 2007 Nov;40(4):287-326. doi: 10.1017/S0033583508004654. Epub 2008 Apr 14. Q Rev Biophys. 2007. PMID: 18405419 Free PMC article. Review.
Cited by
-
Evolutionary adaptation of the protein folding pathway for secretability.EMBO J. 2022 Dec 1;41(23):e111344. doi: 10.15252/embj.2022111344. Epub 2022 Aug 29. EMBO J. 2022. PMID: 36031863 Free PMC article.
-
De novo prediction of protein folding pathways and structure using the principle of sequential stabilization.Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):17442-7. doi: 10.1073/pnas.1209000109. Epub 2012 Oct 8. Proc Natl Acad Sci U S A. 2012. PMID: 23045636 Free PMC article.
-
Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry.Proc Natl Acad Sci U S A. 2013 May 7;110(19):7684-9. doi: 10.1073/pnas.1305887110. Epub 2013 Apr 19. Proc Natl Acad Sci U S A. 2013. PMID: 23603271 Free PMC article.
-
Hydrogen-bond driven loop-closure kinetics in unfolded polypeptide chains.PLoS Comput Biol. 2010 Jan 22;6(1):e1000645. doi: 10.1371/journal.pcbi.1000645. PLoS Comput Biol. 2010. PMID: 20098498 Free PMC article.
-
Multi-layer sequential network analysis improves protein 3D structural classification.Proteins. 2022 Sep;90(9):1721-1731. doi: 10.1002/prot.26349. Epub 2022 May 2. Proteins. 2022. PMID: 35441395 Free PMC article.
References
-
- Anfinsen, C. B. (1973) Science 181, 223-230. - PubMed
-
- Rumbley, J. N., Hoang, L. & Englander, S. W. (2002) Biochemistry 41, 13894-13901. - PubMed
-
- Krishna, M. M. G., Hoang, L., Lin, Y. & Englander, S. W. (2004) Methods 34, 51-64. - PubMed
-
- Connelly, G. P., Bai, Y., Jeng, M. F. & Englander, S. W. (1993) Proteins 17, 87-92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

