Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography
- PMID: 15774580
- PMCID: PMC555690
- DOI: 10.1073/pnas.0409178102
Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography
Abstract
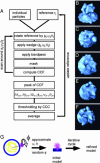
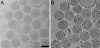
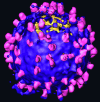
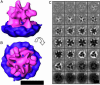
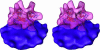
We used cryo-electron tomography in conjunction with single-particle averaging techniques to study the structures of frozen-hydrated envelope glycoprotein (Env) complexes on intact Moloney murine leukemia retrovirus particles. Cryo-electron tomography allows 3D imaging of viruses in toto at a resolution sufficient to locate individual macromolecules, and local averaging of abundant complexes substantially improves the resolution. The averaging of repetitive features in electron tomograms is hampered by a low signal-to-noise ratio and anisotropic resolution, which results from the "missing-wedge" effect. We developed an iterative 3D averaging algorithm that compensates for this effect and used it to determine the trimeric structure of Env to a resolution of 2.7 nm, at which individual domains can be resolved. Strikingly, the 3D reconstruction is shaped like a tripod in which the trimer penetrates the membrane at three distinct locations approximately 4.5 nm apart from one another. The Env reconstruction allows tentative docking of the x-ray crystal structure of the receptor-binding domain. This study thus provides 3D structural information regarding the prefusion conformation of an intact unstained retrovirus surface protein.
Figures

References
-
- Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777-810. - PubMed
-
- Colman, P. M. & Lawrence, M. C. (2003) Nature Rev. Mol. Cell Biol. 4, 309-319. - PubMed
-
- Fass, D., Harrison, S. C. & Kim, P. S. (1996) Nat. Struct. Biol. 3, 465-469. - PubMed
-
- Fass, D., Davey, R. A., Hamson, C. A., Kim, P. S., Cunningham, J. M. & Berger, J. M. (1997) Science 277, 1662-1666. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

