The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy
- PMID: 15774615
- PMCID: PMC1895129
- DOI: 10.1182/blood-2005-01-0204
The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy
Abstract
MLL (ALL-1) chimeric fusions and MLL partial tandem duplications (PTD) may have mechanistically distinct contributions to leukemogenesis. Acute myeloid leukemia (AML) blasts with the t(9;11)(p22; q23) express MLL-AF9 and MLL wild-type (WT) transcripts, while normal karyotype AML blasts with the MLL(PTD/WT) genotype express MLL PTD but not the MLL WT. Silencing of MLL WT in MLL(PTD/WT) blasts was reversed by DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors, and MLL WT induction was associated with selective sensitivity to cell death. Reduction of MLL PTD expression induced MLL WT and reduced blast colony-forming units, supporting opposing functions for MLL PTD and MLL WT whereby the MLL PTD contributes to the leukemic phenotype via a recessive gain-of-function. The coincident suppression of the MLL WT allele with the expression of the MLL PTD allele, along with the functional data presented here, supports the hypothesis that loss of WT MLL function via monoallelic repression contributes to the leukemic phenotype by the remaining mutant allele. These data from primary AML and the pharmacologic reversal of MLL WT silencing associated with a favorable alteration in the threshold for apoptosis suggest that these patients with poor prognosis may benefit from demethylating or histone deacetylase inhibitor therapy, or both.
Figures

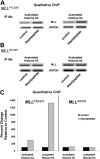
 ), or exon 26 to 27 (not shown) junctions that are common to the MLL WT and MLL PTD transcripts. Primers and probes (▪) were used to detect the MLL PTD-specific exon 11 to 5 fusion or the exon 12 to 5 fusion found in AML cases with either the MLL PTD of exons 5 through 11 or exons 5 through 12, respectively.
), or exon 26 to 27 (not shown) junctions that are common to the MLL WT and MLL PTD transcripts. Primers and probes (▪) were used to detect the MLL PTD-specific exon 11 to 5 fusion or the exon 12 to 5 fusion found in AML cases with either the MLL PTD of exons 5 through 11 or exons 5 through 12, respectively.



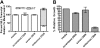
References
-
- Ernst P, Wang J, Korsmeyer SJ. The role of MLL in hematopoiesis and leukemia. Curr Opin Hematol. 2002;9: 282-287. - PubMed
-
- Caligiuri MA, Schichman SA, Strout MP, et al. Molecular rearrangement of the ALL-1 gene in acute myeloid leukemia without cytogenetic evidence of 11q23 chromosomal translocations. Cancer Res. 1994;54: 370-373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

