The Pasteurella multocida nrfE gene is upregulated during infection and is essential for nitrite reduction but not for virulence
- PMID: 15774870
- PMCID: PMC1065219
- DOI: 10.1128/JB.187.7.2278-2285.2005
The Pasteurella multocida nrfE gene is upregulated during infection and is essential for nitrite reduction but not for virulence
Abstract
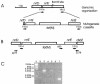
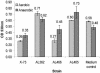
Pasteurella multocida is the causative agent of a range of diseases with economic importance in production animals. Many systems have been employed to identify virulence factors of P. multocida, including in vivo expression technology (IVET), signature-tagged mutagenesis, and whole-genome expression profiling. In a previous study in which IVET was used with P. multocida, nrfE was identified as a gene that is preferentially expressed in vivo. In Escherichia coli, nrfE is part of the formate-dependent nitrite reductase system involved in utilizing available nitrite as an electron accepter during growth under anaerobic conditions. In this study, we constructed an isogenic P. multocida strain that was unable to reduce nitrite under either aerobic or anaerobic conditions, thereby demonstrating that P. multocida nrfE is essential for nitrite reduction. However, the nrfE mutant was still virulent in mice. Real-time reverse transcription-PCR analysis indicated that nrfE was regulated independently of nrfABCD by an independent promoter that is likely to be upregulated in vivo.
Figures

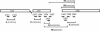
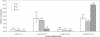

References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
-
- Blattner, F. R., G. Plunkett, 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
-
- Bosch, M., E. Garrido, M. Llagostera, A. M. Perez de Rozas, I. Badiola, and J. Barbe. 2002. Pasteurella multocida exbB, exbD and tonB genes are physically linked but independently transcribed. FEMS Microbiol. Lett. 210:201-218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

