The gusBC genes of Escherichia coli encode a glucuronide transport system
- PMID: 15774881
- PMCID: PMC1065211
- DOI: 10.1128/JB.187.7.2377-2385.2005
The gusBC genes of Escherichia coli encode a glucuronide transport system
Abstract
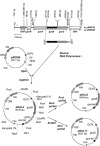
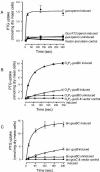
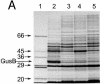
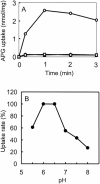
Two genes, gusB and gusC, from a natural fecal isolate of Escherichia coli are shown to encode proteins responsible for transport of beta-glucuronides with synthetic [(14)C]phenyl-1-thio-beta-d-glucuronide as the substrate. These genes are located in the gus operon downstream of the gusA gene on the E. coli genome, and their expression is induced by a variety of beta-d-glucuronides. Measurements of transport in right-side-out subcellular vesicles show the system has the characteristics of secondary active transport energized by the respiration-generated proton motive force. When the genes were cloned together downstream of the tac operator-promoter in the plasmid pTTQ18 expression vector, transport activity was increased considerably with isopropylthiogalactopyranoside as the inducer. Amplified expression of the GusB and GusC proteins enabled visualization and identification by N-terminal sequencing of both proteins, which migrated at ca. 32 kDa and 44 kDa, respectively. Separate expression of the GusB protein showed that it is essential for glucuronide transport and is located in the inner membrane, while the GusC protein does not catalyze transport but assists in an as yet unknown manner and is located in the outer membrane. The output of glucuronides as waste by mammals and uptake for nutrition by gut bacteria or reabsorption by the mammalian host is discussed.
Figures


Similar articles
-
Purification and properties of the Escherichia coli nucleoside transporter NupG, a paradigm for a major facilitator transporter sub-family.Mol Membr Biol. 2004 Sep-Oct;21(5):323-36. doi: 10.1080/09687860400003941. Mol Membr Biol. 2004. PMID: 15513740
-
Studies on the region involved in the transport activity of Escherichia coli TolC by chimeric protein analysis.Microb Pathog. 2007 May-Jun;42(5-6):184-92. doi: 10.1016/j.micpath.2007.01.006. Epub 2007 Feb 8. Microb Pathog. 2007. PMID: 17350794
-
A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli.Mol Microbiol. 2006 Oct;62(1):120-31. doi: 10.1111/j.1365-2958.2006.05326.x. Mol Microbiol. 2006. PMID: 16987175
-
Assembly of membrane-bound respiratory complexes by the Tat protein-transport system.Arch Microbiol. 2002 Aug;178(2):77-84. doi: 10.1007/s00203-002-0434-2. Epub 2002 May 22. Arch Microbiol. 2002. PMID: 12115052 Review.
-
Strategies for Successful Over-Expression of Human Membrane Transport Systems Using Bacterial Hosts: Future Perspectives.Int J Mol Sci. 2022 Mar 30;23(7):3823. doi: 10.3390/ijms23073823. Int J Mol Sci. 2022. PMID: 35409183 Free PMC article. Review.
Cited by
-
Paradoxical Activation of a Type VI Secretion System Phospholipase Effector by Its Cognate Immunity Protein.J Bacteriol. 2023 Jun 27;205(6):e0011323. doi: 10.1128/jb.00113-23. Epub 2023 May 22. J Bacteriol. 2023. PMID: 37212679 Free PMC article.
-
Emergence of a Novel Salmonella enterica Serotype Reading Clonal Group Is Linked to Its Expansion in Commercial Turkey Production, Resulting in Unanticipated Human Illness in North America.mSphere. 2020 Apr 15;5(2):e00056-20. doi: 10.1128/mSphere.00056-20. mSphere. 2020. PMID: 32295868 Free PMC article.
-
MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses.Genome Biol. 2016 Jun 3;17(1):121. doi: 10.1186/s13059-016-0980-6. Genome Biol. 2016. PMID: 27259475 Free PMC article.
-
First Occurrence of the OXA-198 Carbapenemase in Enterobacterales.Antimicrob Agents Chemother. 2020 Mar 24;64(4):e01471-19. doi: 10.1128/AAC.01471-19. Print 2020 Mar 24. Antimicrob Agents Chemother. 2020. PMID: 31932369 Free PMC article.
-
Impediments to enhancement of CPT-11 anticancer activity by E. coli directed beta-glucuronidase therapy.PLoS One. 2015 Feb 17;10(2):e0118028. doi: 10.1371/journal.pone.0118028. eCollection 2015. PLoS One. 2015. PMID: 25688562 Free PMC article.
References
-
- Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
-
- Boos, W., and J. M. Lucht. 1996. Periplasmic binding protein-dependent ABC transporters, p. 1175-1209. In F. C. Neidhardt et al. (ed.) Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
-
- Brissette, J. L., L. Weiner, T. L. Ripmaster, and P. Model. 1991. Characterization and sequence of the Escherichia coli stress-induced psp operon. J. Mol. Biol. 220:35-48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

