Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division
- PMID: 15776021
- PMCID: PMC1299280
- DOI: 10.1038/sj.embor.7400366
Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division
Abstract
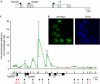
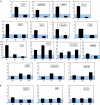
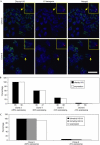
Variant histone H3.3 is incorporated into nucleosomes by a mechanism that does not require DNA replication and has also been implicated as a potential mediator of epigenetic memory of active transcriptional states. In this study, we have used chromatin immunoprecipitation analysis to show that H3.3 is found mainly at the promoters of transcriptionally active genes. We also show that H3.3 combines with H3 acetylation and K4 methylation to form a stable mark that persists during mitosis. Our results suggest that H3.3 is deposited principally through the action of chromatin-remodelling complexes associated with transcriptional initiation, with deposition mediated by RNA polymerase II elongation having only a minor role.
Figures

References
-
- Agalioti T, Chen G, Thanos D (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392 - PubMed
-
- Ahmad K, Henikoff S (2002) The histone variant H3.3 marks active chromatin by repliation-independent nucleosome assembly. Mol Cell 9: 1191–1200 - PubMed
-
- Akhmanova A, Miedema K, Wang Y, van Bruggen M, Berden JH, Moudrianakis EN, Hennig W (1997) The localization of histone H3.3 in germ line chromatin of Drosophila males as established with a histone H3.3specific antiserum. Chromosoma 106: 335–347 - PubMed
-
- Christova R, Oelgeschlager T (2002) Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol 4: 79–82 - PubMed
-
- Henikoff S, Furuyama T, Ahmad K (2004) Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet 20: 320–326 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

