Gapped DNA and cyclization of short DNA fragments
- PMID: 15778443
- PMCID: PMC1305644
- DOI: 10.1529/biophysj.104.055657
Gapped DNA and cyclization of short DNA fragments
Abstract
We use the cyclization of small DNA molecules, approximately 200 bp in length, to study conformational properties of DNA fragments with single-stranded gaps. The approach is extremely sensitive to DNA conformational properties and, being complemented by computations, allows a very accurate determination of the fragment's conformational parameters. Sequence-specific nicking endonucleases are used to create the 4-nt-long gap. We determined the bending rigidity of the single-stranded region in the gapped DNA. We found that the gap of 4 nt in length makes all torsional orientations of DNA ends equally probable. Our results also show that the gap has isotropic bending rigidity. This makes it very attractive to use gapped DNA in the cyclization experiments to determine DNA conformational properties, since the gap eliminates oscillations of the cyclization efficiency with the DNA length. As a result, the number of measurements is greatly reduced in the approach, and the analysis of the data is greatly simplified. We have verified our approach on DNA fragments containing well-characterized intrinsic bends caused by A-tracts. The obtained experimental results and theoretical analysis demonstrate that gapped-DNA cyclization is an exceedingly sensitive and accurate approach for the determination of DNA bending.
Figures



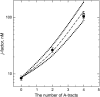
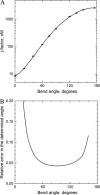
Similar articles
-
Multimerization-cyclization of DNA fragments as a method of conformational analysis.Biophys J. 2000 Nov;79(5):2692-704. doi: 10.1016/S0006-3495(00)76507-6. Biophys J. 2000. PMID: 11053141 Free PMC article.
-
Sequence dependence of DNA bending rigidity.Proc Natl Acad Sci U S A. 2010 Aug 31;107(35):15421-6. doi: 10.1073/pnas.1004809107. Epub 2010 Aug 11. Proc Natl Acad Sci U S A. 2010. PMID: 20702767 Free PMC article.
-
Measurement of the DNA bend angle induced by the catabolite activator protein using Monte Carlo simulation of cyclization kinetics.J Mol Biol. 1998 Feb 13;276(1):287-309. doi: 10.1006/jmbi.1997.1515. J Mol Biol. 1998. PMID: 9514724
-
Strong bending of the DNA double helix.Nucleic Acids Res. 2013 Aug;41(14):6785-92. doi: 10.1093/nar/gkt396. Epub 2013 May 15. Nucleic Acids Res. 2013. PMID: 23677618 Free PMC article. Review.
-
Cruciform structures and functions.Q Rev Biophys. 1996 Dec;29(4):279-307. doi: 10.1017/s0033583500005862. Q Rev Biophys. 1996. PMID: 9080546 Review. No abstract available.
Cited by
-
Nucleosome DNA unwrapping does not affect prototype foamy virus integration efficiency or site selection.PLoS One. 2019 Mar 13;14(3):e0212764. doi: 10.1371/journal.pone.0212764. eCollection 2019. PLoS One. 2019. PMID: 30865665 Free PMC article.
-
Single-molecule analysis of i-motif within self-assembled DNA duplexes and nanocircles.Nucleic Acids Res. 2019 Aug 22;47(14):7199-7212. doi: 10.1093/nar/gkz565. Nucleic Acids Res. 2019. PMID: 31287873 Free PMC article.
-
Effect of magnesium ions and temperature on the sequence-dependent curvature of DNA restriction fragments.J Phys Condens Matter. 2010 Dec 15;22(49):494110. doi: 10.1088/0953-8984/22/49/494110. J Phys Condens Matter. 2010. PMID: 21406776 Free PMC article.
-
When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I.EMBO J. 2006 May 17;25(10):2230-9. doi: 10.1038/sj.emboj.7601104. Epub 2006 Apr 27. EMBO J. 2006. PMID: 16642041 Free PMC article.
-
Motor-like DNA motion due to an ATP-hydrolyzing protein under nanoconfinement.Sci Rep. 2018 Jul 3;8(1):10036. doi: 10.1038/s41598-018-28278-0. Sci Rep. 2018. PMID: 29968756 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

