The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern
- PMID: 15781572
- PMCID: PMC1074360
- DOI: 10.1101/gr.3241705
The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern
Abstract
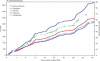
The extent and patterns of linkage disequilibrium (LD) determine the feasibility of association studies to map genes that underlie complex traits. Here we present a comparison of the patterns of LD across four major human populations (African-American, Caucasian, Chinese, and Japanese) with a high-resolution single-nucleotide polymorphism (SNP) map covering almost the entire length of chromosomes 6, 21, and 22. We constructed metric LD maps formulated such that the units measure the extent of useful LD for association mapping. LD reaches almost twice as far in chromosome 6 as in chromosomes 21 or 22, in agreement with their differences in recombination rates. By all measures used, out-of-Africa populations showed over a third more LD than African-Americans, highlighting the role of the population's demography in shaping the patterns of LD. Despite those differences, the long-range contour of the LD maps is remarkably similar across the four populations, presumably reflecting common localization of recombination hot spots. Our results have practical implications for the rational design and selection of SNPs for disease association studies.
Figures


References
-
- Crawford, D.C., Carlson, C.S., Rieder, M.J., Carrington, D.P., Yi, Q., Smith, J.D., Eberle, M.A., Kruglyak, L., and Nickerson, D.A. 2004. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am. J. Hum. Genet. 74: 610-622. - PMC - PubMed
-
- Daly, M.J., Rioux, J.D., Schaffner, S.F., Hudson, T.J., and Lander, E.S. 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29: 229-232. - PubMed
Web site references
-
- http://locus.umdnj.edu/ccr/; Coriell Institute, for details of the NIGMS Human Variation Panels used in this study.
-
- https://myscience.appliedbiosystems.com/; Applied Biosystems' myScience research environment, for the TaqMan Validated SNP Genotyping Assays used in this study.
-
- http://cedar.genetics.soton.ac.uk/pub/PROGRAMS/LDMAP; LDMAP Program.
-
- http://www.allsnps.com/snpbrowser/; SNPbrowser Software, to obtain the genotypes used in this study.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
