Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs
- PMID: 15784600
- PMCID: PMC1087429
- DOI: 10.1128/IAI.73.4.2533-2540.2005
Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs
Abstract
The mechanisms by which Mycobacterium tuberculosis elicits disease are complex, involving a large repertoire of bacterial genes that are required for in vivo growth and survival. To identify such genes, we utilized a high-throughput microarray detection method to rapidly screen hundreds of unique, genotypically defined transposon mutants for in vivo survival with a high degree of specificity and sensitivity. Thirty-one M. tuberculosis genes were found to be required for in vivo survival in mouse lungs. These genes are involved in a broad range of activities, including metabolism, cell wall functions, and regulation. Our screen included 11 of the 12 known members of the mycobacterial membrane protein (mmpL) family genes, and mutation of 6 of these genes-mmpL4, mmpL5, mmpL7, mmpL8, mmpL10, and mmpL11-severely compromised the ability of the mutants to multiply in mouse lungs. Most of the 31 genes are conserved in other pathogenic mycobacteria, including M. leprae and M. bovis, suggesting that a core of basic in vivo survival mechanisms may be highly conserved despite the divergent human pathology caused by members of the mycobacterial genus. Of the 31 genes reported here, 17 have not been previously described to be involved in in vivo growth and survival of M. tuberculosis.
Figures


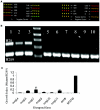
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols and molecular biology. John Wiley & Sons, New York, N.Y.
-
- Avarbock, D., J. Salem, L. S. Li, Z. M. Wang, and H. Rubin. 1999. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233:261-269. - PubMed
-
- Camacho, L. R., P. Constant, C. Raynaud, M. A. Laneelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis: evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. - PubMed
-
- Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

