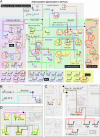
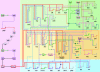
Fig. 2.
The dorsal–ventral GRN in Drosophila. The overall presentation is similar to that in Fig. 1. The diagram represents regulatory inputs and outputs for 46 genes expressed in the early embryo, from 2 to 5 h after fertilization. During this 3-h window, the syncytial embryo undergoes cellularization, mesoderm invagination, and the rapid phase of germband elongation. The color coding, from bottom to top, represents the three primary embryonic tissues as follows: mesoderm (Bottom, blue), ventral neurogenic ectoderm (Middle, yellow), and dorsal neurogenic ectoderm plus dorsal ectoderm (Top, yellow). The light shading to the left of the diagram represents syncytial stages, between 2 and 3 h after fertilization. The darker shading to the right represents cellularized embryos undergoing gastrulation. Dorsal–ventral patterning is initiated by the graded distribution of the Dorsal transcription factor. Peak levels of Dorsal enter nuclei in ventral (bottom) regions of the embryo, intermediate levels in lateral regions that form the ventral neurogenic ectoderm, and low levels in the dorsal neurogenic ectoderm. This Dorsal nuclear gradient is formed by the differential activation of the Toll signaling pathway (35), which in turn depends on the localized transcription of pipe in ventral follicle cells of the egg chamber (57). The pipe gene is probably repressed by EGF signaling, which is restricted to dorsal follicle cells because of the asymmetric position of the oocyte nucleus (58). Localized transcription of pipe in ventral follicle cells leads to a serine protease cascade on the ventral surface of the growing oocyte (ndl, gd, snk, and ea) that cleaves an inactive precursor form of the Spatzle (spz) ligand (59). The active ligand is thought to be deposited in a graded fashion along the ventral and lateral surface of the unfertilized egg. After fertilization, the Spz gradient leads to the Dorsal nuclear gradient within the syncytial embryo. High levels of Dorsal activate several genes in ventral regions that constitute the presumptive mesoderm, including twist (twi), snail (sna), NF-YC (a specialized component of the general NF-Y CCAAT binding complex), the FGF Heartless receptor (Htl), and Heartbroken (Hbr; also called Dof and Stumps), which transduces FGF signaling within the cell (30, 36). Twi is an activator that works in concert with Dorsal to activate sna expression in the mesoderm (9), and there is evidence that Twi also helps activate htl and hbr (36). Dorsal, Twi, and Sna regulate a large number of genes during the syncytial phases of dorsal–ventral patterning, including brk, vnd, rho, and vn, which are selectively activated in ventral regions of the neurogenic ectoderm (60). Dorsal and Twi work in a synergistic fashion to activate these genes, whereas the Sna repressor excludes their expression from the ventral mesoderm. Low levels of the Dorsal gradient activate short gastrulation (sog) and thisbe (ths) throughout the neurogenic ectoderm, in both dorsal and ventral regions (9, 30). Both genes encode secreted signaling molecules; Sog inhibits Dpp signaling (61), whereas Ths is related to FGF8 and activates FGF signaling in the dorsal mesoderm during gastrulation (see below). Low levels of Dorsal also repress tolloid (tld), zerknullt (zen), and decapentaplegic (dpp), which are required for the patterning of the dorsal ectoderm after cellularization (9). Definitive tissues begin to arise from each of the generic embryonic territories at the onset of gastrulation. The shading highlights the tinman (tin) and even-skipped (eve) genes, which gives rise to derivatives of the dorsal mesoderm such as visceral and cardiac muscles (31). eve is activated by Twi, Tin, Ets-containing transcription factors induced by FGF signaling, and Smad transcription factors induced by Dpp signaling after the internal dorsal mesoderm comes into contact with the dorsal ectoderm after gastrulation (31, 62). The shading in the central neurogenic ectoderm highlights a positive feedback system that is coordinated by the regulatory gene sim. sim is activated by Dorsal, Twi, and Su(H), the transcriptional effector of Notch signaling (44, 45). An unknown Notch signal emanating from the mesoderm induces sim expression in the ventral-most row of cells in the neurogenic ectoderm. Sim activates several components of the EGF signaling pathway, including rho, star, and spitz (–49). Rho and Star are required for the processing of the Spitz ligand (63), which activates a ubiquitous EGF receptor (egfr). Activation of EGF signaling leads to the induction of pointed p1 (pnt) expression, which activates orthodenticle (otd) in the ventral midline (51, 52). EGF signaling and pnt either directly or indirectly maintain the expression of several genes in the neurogenic ectoderm that were previously activated by Dorsal plus Twi, including ind and vnd, which encode regulatory proteins that pattern the future ventral nerve cord (53, 55). Sim also participates in the activation of slit (sli), which encodes a signaling molecule required for the proper organization of the neurons that comprise the nerve cord (50). Finally, the shading on top (right) highlights the differentiation of two derivatives of the dorsal ectoderm: the dorsal epidermis and amnioserosa. A Dpp activity gradient is created in the dorsal ectoderm from the combined action of the Sog inhibitor emanating from the neurogenic ectoderm and the Tld protease, which releases Dpp from Sog at the dorsal midline (61). Dpp works together with a ubiquitous bone morphogenetic protein (BMP) signaling molecule called Screw (Scw). Peak levels of Dpp and Scw signaling at the dorsal midline lead to the phosphorylation and nuclear transport of two Smad transcription factors, Mad and Medea (med) (64). Mad and Medea, along with the Zen homeodomain regulator, activate a number of genes required for the differentiation and function of the amnioserosa, including hindsight (hnt) and Doc (a Tbx6 transcription factor) (65). Lower levels of Dpp plus Scw signaling activate a number of regulatory genes throughout the dorsal ectoderm, including tailup (tup), u-shaped (ush), pannier (pnr), and schnurri (shn) (66). These genes respond to lower levels of Mad plus Medea, or as drawn in the diagram, respond solely to a particular activator complex containing Medea. Shn functions as a repressor that maintains the boundary between the neurogenic ectoderm and dorsal ectoderm by repressing brk (43) and neurogenic genes such as msh, which is expressed in the dorsal-most regions of the neurogenic ectoderm (53).