Preferences for the selection of unique tRNA primers revealed from analysis of HIV-1 replication in peripheral blood mononuclear cells
- PMID: 15790410
- PMCID: PMC1084362
- DOI: 10.1186/1742-4690-2-21
Preferences for the selection of unique tRNA primers revealed from analysis of HIV-1 replication in peripheral blood mononuclear cells
Abstract
Background: All human immunodeficiency virus (HIV-1) uses a host tRNALys,3 as the primer for reverse transcription. The tRNALys,3 is bound to a region on the HIV-1 genome, the primer-binding site (PBS), that is complementary to the 18 terminal nucleotides of tRNALys,3. How HIV-1 selects the tRNA from the intracellular milieu is unresolved.
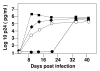
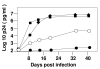
Results: HIV-1 tRNA primer selection has been investigated using viruses in which the primer-binding site (PBS) and a sequence within U5 were altered so as to be complementary to tRNAMet, tRNAPro or tRNAIle. Analysis of the replication of these viruses in human peripheral blood mononuclear cells (PBMC) revealed preferences for the selection of certain tRNAs. HIV-1 with the PBS altered to be complementary to tRNAMet, with and without the additional mutation in U5 to be complementary to the anticodon of tRNAMet, stably maintains the PBS complementary to tRNAMet following extended in vitro culture in PBMC. In contrast, viruses with either the PBS or PBS and U5 mutated to be complementary to tRNAIle were unstable during in vitro replication in PBMC and reverted to utilize tRNALys,3. Viruses with the PBS altered to be complementary to tRNAPro replicated in PBMC but reverted to use tRNALys,3; viruses with mutations in both the U5 and PBS complementary to tRNAPro maintained this PBS, yet replicated poorly in PBMC.
Conclusion: The results of these studies demonstrate that HIV-1 has preferences for selection of certain tRNAs for high-level replication in PBMC.
Figures


Similar articles
-
Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription.Virology. 1996 Aug 15;222(2):401-14. doi: 10.1006/viro.1996.0437. Virology. 1996. PMID: 8806524
-
HIV type 1 that select tRNA(His) or tRNA(Lys1,2) as primers for reverse transcription exhibit different infectivities in peripheral blood mononuclear cells.AIDS Res Hum Retroviruses. 2004 Apr;20(4):373-81. doi: 10.1089/088922204323048122. AIDS Res Hum Retroviruses. 2004. PMID: 15157356
-
The G490E mutation in reverse transcriptase does not impact tRNA primer selection by HIV-1 with altered PBS and A-loop.Virology. 2006 Sep 1;352(2):380-9. doi: 10.1016/j.virol.2006.04.019. Epub 2006 Jun 15. Virology. 2006. PMID: 16781756
-
HIV-1 reverse transcription initiation: a potential target for novel antivirals?Virus Res. 2008 Jun;134(1-2):4-18. doi: 10.1016/j.virusres.2007.12.009. Epub 2008 Feb 6. Virus Res. 2008. PMID: 18255184 Review.
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
Cited by
-
Specificity of phage display selected peptides for modified anticodon stem and loop domains of tRNA.Protein J. 2007 Jan;26(1):61-73. doi: 10.1007/s10930-006-9046-z. Protein J. 2007. PMID: 17237992
-
HIV-1 Subtypes and 5'LTR-Leader Sequence Variants Correlate with Seroconversion Status in Pumwani Sex Worker Cohort.Viruses. 2017 Dec 23;10(1):4. doi: 10.3390/v10010004. Viruses. 2017. PMID: 29295533 Free PMC article.
-
HIV-1 designed to use different tRNAGln isoacceptors prefers to select tRNAThr for replication.Virol J. 2006 Sep 26;3:80. doi: 10.1186/1743-422X-3-80. Virol J. 2006. PMID: 17002807 Free PMC article.
-
HIV gene expression from intact proviruses positioned in bacterial artificial chromosomes at integration sites previously identified in latently infected T cells.Virology. 2011 Feb 5;410(1):151-60. doi: 10.1016/j.virol.2010.11.001. Epub 2010 Nov 27. Virology. 2011. PMID: 21115184 Free PMC article.
-
Analysis of the replication of HIV-1 forced to use tRNAMet(i) supports a link between primer selection, translation and encapsidation.Retrovirology. 2007 Feb 2;4:10. doi: 10.1186/1742-4690-4-10. Retrovirology. 2007. PMID: 17274824 Free PMC article.
References
-
- Harada F, Sawyer RC, Dahlberg JE. A primer ribonucleic acid for initiation of in vitro Rous sarcoma virus deoxyribonucleic acid synthesis. J Biol Chem. 1975;250:3487–3497. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

