Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
- PMID: 15791207
- PMCID: PMC1142568
- DOI: 10.1038/sj.emboj.7600632
Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
Abstract
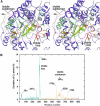
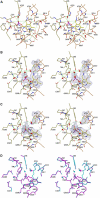
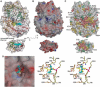
The crystal structure of the P-protein of the glycine cleavage system from Thermus thermophilus HB8 has been determined. This is the first reported crystal structure of a P-protein, and it reveals that P-proteins do not involve the alpha(2)-type active dimer universally observed in the evolutionarily related pyridoxal 5'-phosphate (PLP)-dependent enzymes. Instead, novel alphabeta-type dimers associate to form an alpha(2)beta(2) tetramer, where the alpha- and beta-subunits are structurally similar and appear to have arisen by gene duplication and subsequent divergence with a loss of one active site. The binding of PLP to the apoenzyme induces large open-closed conformational changes, with residues moving up to 13.5 A. The structure of the complex formed by the holoenzyme bound to an inhibitor, (aminooxy)acetate, suggests residues that may be responsible for substrate recognition. The molecular surface around the lipoamide-binding channel shows conservation of positively charged residues, which are possibly involved in complex formation with the H-protein. These results provide insights into the molecular basis of nonketotic hyperglycinemia.
Figures




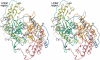
References
-
- Adachi S, Oguchi T, Tanida H, Park SY, Shimizu H, Miyatake H, Kamiya N, Shiro Y, Inoue Y, Ueki T, Iizuka T (2001) The RIKEN structural biology beamline II (BL44B2) at the SPring-8. Nucl Instrum Methods A 467: 711–714
-
- Alexeev D, Alexeeva M, Baxter RL, Campopiano DJ, Webster SP, Sawyer L (1998) The crystal structure of 8-amino-7-oxononanoate synthase: a bacterial PLP-dependent, acyl-CoA-condensing enzyme. J Mol Biol 284: 401–419 - PubMed
-
- Applegarth DA, Toone JR (2001) Nonketotic hyperglycinemia (glycine encephalopathy): laboratory diagnosis. Mol Genet Metab 74: 139–146 - PubMed
-
- Bertoldi M, Gonsalvi M, Contestabile R, Voltattorni CB (2002) Mutation of tyrosine 332 to phenylalanine converts dopa decarboxylase into a decarboxylation-dependent oxidative deaminase. J Biol Chem 277: 36357–36362 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

