LPS induces the interaction of a transcription factor, LPS-induced TNF-alpha factor, and STAT6(B) with effects on multiple cytokines
- PMID: 15793005
- PMCID: PMC555602
- DOI: 10.1073/pnas.0501159102
LPS induces the interaction of a transcription factor, LPS-induced TNF-alpha factor, and STAT6(B) with effects on multiple cytokines
Abstract
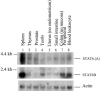
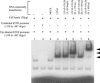
TNF-alpha is a pivotal cytokine whose overproduction can be lethal. Previously, we identified a transcription factor, LPS-induced TNF-alpha factor (LITAF), that regulates TNF-alpha transcription. We now report the discovery and characterization of a regulatory cofactor that we call signal transducer and activator of transcription (STAT) 6(B) because of its considerable homology to STAT6 [here referred to as STAT6(A)]. The STAT6(B) gene expression was found to be activated by LPS. Furthermore, we show that cotransfection of STAT6(B) and LITAF induces an interaction between the two proteins, consequently forming a complex that subsequently translocates into the nucleus and up-regulates the transcription of cytokines. The effect of the complex on a panel of cytokines was tested. In addition, the specific role of LITAF in this complex was established with experiments, including RNA interference technology. Overall, these findings describe roles for LITAF, STAT6(B), and the LITAF-STAT6(B) complex in the regulation of inflammatory cytokines in response to LPS stimulation in mammalian cells.
Figures





Similar articles
-
LPS-induced TNF-α factor mediates pro-inflammatory and pro-fibrogenic pattern in non-alcoholic fatty liver disease.Oncotarget. 2015 Dec 8;6(39):41434-52. doi: 10.18632/oncotarget.5163. Oncotarget. 2015. PMID: 26573228 Free PMC article.
-
LPS-induced TNF-alpha factor (LITAF)-deficient mice express reduced LPS-induced cytokine: Evidence for LITAF-dependent LPS signaling pathways.Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13777-82. doi: 10.1073/pnas.0605988103. Epub 2006 Sep 5. Proc Natl Acad Sci U S A. 2006. PMID: 16954198 Free PMC article.
-
Novel regulation of CCL2 gene expression by murine LITAF and STAT6B.PLoS One. 2011;6(9):e25083. doi: 10.1371/journal.pone.0025083. Epub 2011 Sep 28. PLoS One. 2011. PMID: 21980379 Free PMC article.
-
Lipopolysaccharide-induced tumor necrosis factor-α factor enhances inflammation and is associated with cancer (Review).Mol Med Rep. 2015 Nov;12(5):6399-404. doi: 10.3892/mmr.2015.4243. Epub 2015 Aug 25. Mol Med Rep. 2015. PMID: 26324337 Review.
-
The biology of Stat4 and Stat6.Oncogene. 2000 May 15;19(21):2577-84. doi: 10.1038/sj.onc.1203485. Oncogene. 2000. PMID: 10851056 Review.
Cited by
-
Intestinal Microbiomics and Metabolomics Insights into the Hepatoprotective Effects of Lactobacillus paracasei CCFM1222 Against the Acute Liver Injury in Mice.Probiotics Antimicrob Proteins. 2023 Oct;15(5):1063-1077. doi: 10.1007/s12602-022-09986-6. Epub 2022 Sep 2. Probiotics Antimicrob Proteins. 2023. PMID: 36056292
-
LITAF, a BCL6 target gene, regulates autophagy in mature B-cell lymphomas.Br J Haematol. 2013 Sep;162(5):621-30. doi: 10.1111/bjh.12440. Epub 2013 Jun 25. Br J Haematol. 2013. PMID: 23795761 Free PMC article.
-
Lactobacillus paracasei CCFM1223 Protects against Lipopolysaccharide-Induced Acute Liver Injury in Mice by Regulating the "Gut-Liver" Axis.Microorganisms. 2022 Jun 30;10(7):1321. doi: 10.3390/microorganisms10071321. Microorganisms. 2022. PMID: 35889040 Free PMC article.
-
Curcumin prevents human dendritic cell response to immune stimulants.Biochem Biophys Res Commun. 2008 Sep 26;374(3):431-6. doi: 10.1016/j.bbrc.2008.07.051. Epub 2008 Jul 17. Biochem Biophys Res Commun. 2008. PMID: 18639521 Free PMC article.
-
Highly conserved transcriptional responses to mechanical ventilation of the lung.Physiol Genomics. 2010 Aug;42(3):384-96. doi: 10.1152/physiolgenomics.00117.2009. Epub 2010 May 11. Physiol Genomics. 2010. PMID: 20460603 Free PMC article.
References
-
- Beutler, B. & Cerami, A. (1989) Annu. Rev. Immunol. 7, 625–655. - PubMed
-
- Kramer, B., Wiegmann, K. & Kronke, M. (1995) J. Biol. Chem. 270, 6577–6583. - PubMed
-
- Lee, M. H, Park, J., Chung, S. W., Kang, B. Y., Kim, S. H. & Kim, T. S. (2004) Int. Arch. Allergy Immunol. 134, 213–222. - PubMed
-
- Vitiello, M., D'Isanto, M., Galdiero, M., Raieta, K., Tortora, A., Rotondo, P., Peluso, L. & Galdiero, M. (2004) Cytokine 27, 15–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

