The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila
- PMID: 15793007
- PMCID: PMC555997
- DOI: 10.1073/pnas.0500373102
The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila
Abstract
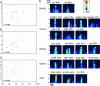
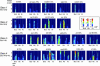
The maternal morphogen Bicoid (Bcd) is distributed in an embryonic gradient that is critical for patterning the anterior-posterior (AP) body plan in Drosophila. Previous work identified several target genes that respond directly to Bcd-dependent activation. Positioning of these targets along the AP axis is thought to be controlled by cis-regulatory modules (CRMs) that contain clusters of Bcd-binding sites of different "strengths." Here we use a combination of Bcd-site cluster analysis and evolutionary conservation to predict Bcd-dependent CRMs. We tested 14 predicted CRMs by in vivo reporter gene assays; 11 show Bcd-dependent activation, which brings the total number of known Bcd target elements to 21. Some CRMs drive expression patterns that are restricted to the most anterior part of the embryo, whereas others extend into middle and posterior regions. However, we do not detect a strong correlation between AP position of target gene expression and the strength of Bcd site clusters alone. Rather, we find that binding sites for other activators, including Hunchback and Caudal correlate with CRM expression in middle and posterior body regions. Also, many Bcd-dependent CRMs contain clusters of sites for the gap protein Kruppel, which may limit the posterior extent of activation by the Bcd gradient. We propose that the key design principle in AP patterning is the differential integration of positive and negative transcriptional information at the level of individual CRMs for each target gene.
Figures


Similar articles
-
Uncovering a dynamic feature of the transcriptional regulatory network for anterior-posterior patterning in the Drosophila embryo.PLoS One. 2013 Apr 30;8(4):e62641. doi: 10.1371/journal.pone.0062641. Print 2013. PLoS One. 2013. PMID: 23646132 Free PMC article.
-
Anterior-posterior positional information in the absence of a strong Bicoid gradient.Proc Natl Acad Sci U S A. 2009 Mar 10;106(10):3823-8. doi: 10.1073/pnas.0807878105. Epub 2009 Feb 23. Proc Natl Acad Sci U S A. 2009. PMID: 19237583 Free PMC article.
-
A feed-forward relay integrates the regulatory activities of Bicoid and Orthodenticle via sequential binding to suboptimal sites.Genes Dev. 2018 May 1;32(9-10):723-736. doi: 10.1101/gad.311985.118. Epub 2018 May 15. Genes Dev. 2018. PMID: 29764918 Free PMC article.
-
How to get ahead: the origin, evolution and function of bicoid.Bioessays. 2005 Sep;27(9):904-13. doi: 10.1002/bies.20285. Bioessays. 2005. PMID: 16108065 Review.
-
A matter of time: Formation and interpretation of the Bicoid morphogen gradient.Curr Top Dev Biol. 2020;137:79-117. doi: 10.1016/bs.ctdb.2019.11.016. Epub 2019 Dec 27. Curr Top Dev Biol. 2020. PMID: 32143754 Review.
Cited by
-
Pattern formation by graded and uniform signals in the early Drosophila embryo.Biophys J. 2012 Feb 8;102(3):427-33. doi: 10.1016/j.bpj.2011.12.042. Epub 2012 Feb 7. Biophys J. 2012. PMID: 22325264 Free PMC article.
-
The gap gene network.Cell Mol Life Sci. 2011 Jan;68(2):243-74. doi: 10.1007/s00018-010-0536-y. Epub 2010 Oct 8. Cell Mol Life Sci. 2011. PMID: 20927566 Free PMC article. Review.
-
Two coacting shadow enhancers regulate twin of eyeless expression during early Drosophila development.Genetics. 2025 Jan 8;229(1):1-43. doi: 10.1093/genetics/iyae176. Genetics. 2025. PMID: 39607769 Free PMC article.
-
A mechanism for hunchback promoters to readout morphogenetic positional information in less than a minute.Elife. 2020 Jul 29;9:e49758. doi: 10.7554/eLife.49758. Elife. 2020. PMID: 32723476 Free PMC article.
-
cis-Decoder discovers constellations of conserved DNA sequences shared among tissue-specific enhancers.Genome Biol. 2007;8(5):R75. doi: 10.1186/gb-2007-8-5-r75. Genome Biol. 2007. PMID: 17490485 Free PMC article.
References
-
- Driever, W. & Nusslein-Volhard, C. (1988) Cell 54, 83–93. - PubMed
-
- Driever, W. & Nusslein-Volhard, C. (1988) Cell 54, 95–104. - PubMed
-
- Ephrussi, A. & St Johnston, D. (2004) Cell 116, 143–152. - PubMed
-
- Driever, W. (1993) in The Development of Drosophila melanogaster, eds. Bate, M. & Martinez-Arias, A. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 301–324.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

