Development of a cell-based high-throughput specificity screen using a hepatitis C virus-bovine viral diarrhea virus dual replicon assay
- PMID: 15793110
- PMCID: PMC1068622
- DOI: 10.1128/AAC.49.4.1346-1353.2005
Development of a cell-based high-throughput specificity screen using a hepatitis C virus-bovine viral diarrhea virus dual replicon assay
Abstract
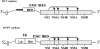
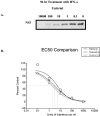
The hepatitis C virus (HCV) replicon is a unique system for the development of a high-throughput screen (HTS), since the analysis of inhibitors requires the quantification of a decrease in a steady-state level of HCV RNA. HCV replicon replication is dependent on host cell factors, and any toxic effects may have a significant impact on HCV replicon replication. Therefore, determining the antiviral specificity of compounds presents a challenge for the identification of specific HCV inhibitors. Here we report the development of an HCV/bovine viral diarrhea virus (BVDV) dual replicon assay suitable for HTS to address these issues. The HCV reporter enzyme is the endogenous NS3 protease contained within the HCV genome, while the BVDV reporter enzyme is a luciferase enzyme engineered into the BVDV genome. The HTS uses a mixture of HCV and BVDV replicon cell lines placed in the same well of a 96-well plate and isolated in the same cell backgrounds (Huh-7). The format consists of three separate but compatible assays: the first quantitates the amount of cytotoxicity based upon the conversion of Alamar blue dye via cellular enzymes, while the second indirectly quantitates HCV replicon replication through measurement of the amount of NS3 protease activity present. The final assay measures the amount of luciferase activity present from the BVDV replicon cells, as an indicator of the specificity of the test compounds. This HCV/BVDV dual replicon assay provides a reliable format to determine the potency and specificity of HCV replicon inhibitors.
Figures

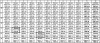

References
-
- Ahmed, S., J. Gopal, Jr., and J. Walsh. 1994. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: An alternative to H3-thymidine incorporation assay. J. Immunol. Methods 170:211-224. - PubMed
-
- Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 52:1-17. - PubMed
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

