Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG
- PMID: 15793135
- PMCID: PMC1068620
- DOI: 10.1128/AAC.49.4.1529-1541.2005
Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG
Abstract
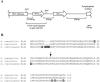
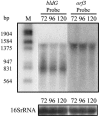
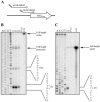
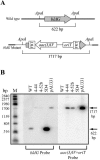
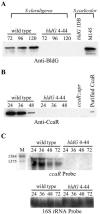
In Streptomyces coelicolor, bldG encodes a putative anti-anti-sigma factor that regulates both aerial hypha formation and antibiotic production, and a downstream transcriptionally linked open reading frame (orf3) encodes a putative anti-sigma factor protein. A cloned DNA fragment from Streptomyces clavuligerus contained an open reading frame that encoded a protein showing 92% identity to the S. coelicolor BldG protein and 91% identity to the BldG ortholog in Streptomyces avermitilis. Sequencing of the region downstream of bldG in S. clavuligerus revealed the presence of an open reading frame encoding a protein showing 72 and 69% identity to the ORF3 proteins in S. coelicolor and S. avermitilis, respectively. Northern analysis indicated that, as in S. coelicolor, the S. clavuligerus bldG gene is expressed as both a monocistronic and a polycistronic transcript, the latter including the downstream orf3 gene. High-resolution S1 nuclease mapping of S. clavuligerus bldG transcripts revealed the presence of three bldG-specific promoters, and analysis of expression of a bldGp-egfp reporter indicated that the bldG promoter is active at various stages of development and in both substrate and aerial hyphae. A bldG null mutant was defective in both morphological differentiation and in the production of secondary metabolites, such as cephamycin C, clavulanic acid, and the 5S clavams. This inability to produce cephamycin C and clavulanic acid was due to the absence of the CcaR transcriptional regulator, which controls the expression of biosynthetic genes for both secondary metabolites as well as the expression of a second regulator of clavulanic acid biosynthesis, ClaR. This makes bldG the first regulatory protein identified in S. clavuligerus that functions upstream of CcaR and ClaR in a regulatory cascade to control secondary metabolite production.
Figures

References
-
- Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. - PubMed
-
- Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195-205. - PubMed
-
- Bailey, C. R., M. J. Butler, I. D. Normansell, R. T. Rowlands, and D. J. Winstanley. 1984. Cloning a Streptomyces clavuligerus genetic locus involved in clavulanic acid biosynthesis. Bio/Technology 2:808-811.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

