Requirements at the 3' end of the sindbis virus genome for efficient synthesis of minus-strand RNA
- PMID: 15795249
- PMCID: PMC1069581
- DOI: 10.1128/JVI.79.8.4630-4639.2005
Requirements at the 3' end of the sindbis virus genome for efficient synthesis of minus-strand RNA
Abstract
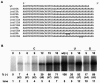
The 3'-untranslated region of the Sindbis virus genome is 0.3 kb in length with a 19-nucleotide conserved sequence element (3' CSE) immediately preceding the 3'-poly(A) tail. The 3' CSE and poly(A) tail have been assumed to constitute the core promoter for minus-strand RNA synthesis during genome replication; however, their involvement in this process has not been formally demonstrated. Utilizing both in vitro and in vivo analyses, we have examined the role of these elements in the initiation of minus-strand RNA synthesis. The major findings of this study with regard to efficient minus-strand RNA synthesis are the following: (i) the wild-type 3' CSE and the poly(A) tail are required, (ii) the poly(A) tail must be a minimum of 11 to 12 residues in length and immediately follow the 3' CSE, (iii) deletion or substitution of the 3' 13 nucleotides of the 3' CSE severely inhibits minus-strand RNA synthesis, (iv) templates possessing non-wild-type 3' sequences previously demonstrated to support virus replication do not program efficient RNA synthesis, and (v) insertion of uridylate residues between the poly(A) tail and a non-wild-type 3' sequence can restore promoter function to a limited extent. This study shows that the optimal structure of the 3' component of the minus-strand promoter is the wild-type 3' CSE followed a poly(A) tail of at least 11 residues. Our findings also show that insertion of nontemplated bases can restore function to an inactive promoter.
Figures

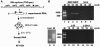

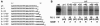
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

