CD4+ T-cell responses to Epstein-Barr virus (EBV) latent-cycle antigens and the recognition of EBV-transformed lymphoblastoid cell lines
- PMID: 15795275
- PMCID: PMC1069546
- DOI: 10.1128/JVI.79.8.4896-4907.2005
CD4+ T-cell responses to Epstein-Barr virus (EBV) latent-cycle antigens and the recognition of EBV-transformed lymphoblastoid cell lines
Abstract
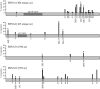
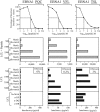
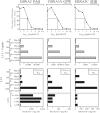
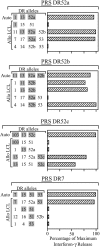
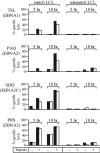
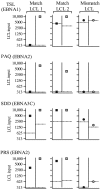
There is considerable interest in the potential of Epstein-Barr virus (EBV) latent antigen-specific CD4+ T cells to act as direct effectors controlling EBV-induced B lymphoproliferations. Such activity would require direct CD4+ T-cell recognition of latently infected cells through epitopes derived from endogenously expressed viral proteins and presented on the target cell surface in association with HLA class II molecules. It is therefore important to know how often these conditions are met. Here we provide CD4+ epitope maps for four EBV nuclear antigens, EBNA1, -2, -3A, and -3C, and establish CD4+ T-cell clones against 12 representative epitopes. For each epitope we identify the relevant HLA class II restricting allele and determine the efficiency with which epitope-specific effectors recognize the autologous EBV-transformed B-lymphoblastoid cell line (LCL). The level of recognition measured by gamma interferon release was consistent among clones to the same epitope but varied between epitopes, with values ranging from 0 to 35% of the maximum seen against the epitope peptide-loaded LCL. These epitope-specific differences, also apparent in short-term cytotoxicity and longer-term outgrowth assays on LCL targets, did not relate to the identity of the source antigen and could not be explained by the different functional avidities of the CD4+ clones; rather, they appeared to reflect different levels of epitope display at the LCL surface. Thus, while CD4+ T-cell responses are detectable against many epitopes in EBV latent proteins, only a minority of these responses are likely to have therapeutic potential as effectors directly recognizing latently infected target cells.
Figures


References
-
- Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. - PubMed
-
- Chen, M., M. Shirai, Z. Liu, T. Arichi, H. Takahashi, and M. Nishioka. 1998. Efficient class II major histocompatibility complex presentation of endogenously synthesized hepatitis C virus core protein by Epstein-Barr virus-transformed B-lymphoblastoid cell lines to CD4+ T cells. J. Virol. 72:8301-8308. - PMC - PubMed
-
- Crotzer, V. L., R. E. Christian, J. M. Brooks, J. Shabanowitz, R. E. Settlage, J. A. Marto, F. M. White, A. B. Rickinson, D. F. Hunt, and V. H. Engelhard. 2000. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. J. Immunol. 164:6120-6129. - PubMed
-
- Greenspan, J. S., D. Greenspan, E.T. Lennette, D. I. Abrams, M. A. Conant, V. Petersen, and U. K. Freese. 1985. Replication of Epstein-Barr virus within the epithelial cells of oral “hairy” leukoplakia, an AIDS-associated lesion. N. Engl. J. Med. 313:1564-1571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

