In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA
- PMID: 15795303
- PMCID: PMC1069538
- DOI: 10.1128/JVI.79.8.5203-5210.2005
In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA
Abstract
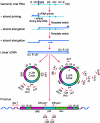
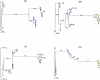
Current regimens for the management of human immunodeficiency virus type 1 (HIV-1) infection suppress plasma viremia to below detectable levels for prolonged intervals. Nevertheless, there is a rapid resumption in plasma viremia if therapy is interrupted. Attempts to characterize the extent of viral replication under conditions of potent suppression and undetectable plasma viremia have been hampered by a lack of convenient assays that can distinguish latent from ongoing viral replication. Using episomal viral cDNA as a surrogate for ongoing replication, we previously presented evidence that viral replication persists in the majority of infected individuals with a sustained aviremic status. The labile nature of viral episomes and hence their validity as surrogate markers of ongoing replication in individuals with long-term-suppressed HIV-1 infection have been analyzed in short-term in vitro experiments with conflicting results. Since these in vitro experiments do not shed light on the long-term in vivo dynamics of episomal cDNA or recapitulate the natural targets of infection in vivo, we have analyzed the dynamics of episomal cDNA turnover in vivo by following the emergence of an M184V polymorphism in plasma viral RNA, in episomal cDNA, and in proviral DNA in patients on suboptimal therapies. We demonstrate that during acquisition of drug resistance, wild-type episomal cDNAs are replaced by M184V-harboring episomes. Importantly, a complete replacement of wild-type episomes with M184V-containing episomes occurred while proviruses remained wild type. This indicates that episomal cDNAs are turned over by degradation rather than through death or tissue redistribution of the infected cell itself. Therefore, evolution of episomal viral cDNAs is a valid surrogate of ongoing viral replication in HIV-1-infected individuals.
Figures


References
-
- Bi, X., H. Gatanaga, S. Ida, K. Tsuchiya, S. Matsuoka-Aizawa, S. Kimura, and S. Oka. 2003. Emergence of protease inhibitor resistance-associated mutations in plasma HIV-1 precedes that in proviruses of peripheral blood mononuclear cells by more than a year. J. Acquir. Immune Defic. Syndr. 34:1-6. - PubMed
-
- Brooks, D. G., D. H. Hamer, P. A. Arlen, L. Gao, G. Bristol, C. M. Kitchen, E. A. Berger, and J. A. Zack. 2003. Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19:413-423. - PubMed
-
- Bushman, F. 2003. Measuring covert HIV replication during HAART: the abundance of 2-LTR circles is not a reliable marker. AIDS 17:749-750. - PubMed
-
- Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

