The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress
- PMID: 15795314
- PMCID: PMC2171834
- DOI: 10.1083/jcb.200412076
The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress
Abstract
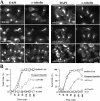
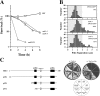
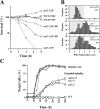
The budding yeast S phase checkpoint responds to hydroxyurea-induced nucleotide depletion by preventing replication fork collapse and the segregation of unreplicated chromosomes. Although the block to chromosome segregation has been thought to occur by inhibiting anaphase, we show checkpoint-defective rad53 mutants undergo cycles of spindle extension and collapse after hydroxyurea treatment that are distinct from anaphase cells. Furthermore, chromatid cohesion, whose dissolution triggers anaphase, is dispensable for S phase checkpoint arrest. Kinetochore-spindle attachments are required to prevent spindle extension during replication blocks, and chromosomes with two centromeres or an origin of replication juxtaposed to a centromere rescue the rad53 checkpoint defect. These observations suggest that checkpoint signaling is required to generate an inward force involved in maintaining preanaphase spindle integrity during DNA replication distress. We propose that by promoting replication fork integrity under these conditions Rad53 ensures centromere duplication. Replicating chromosomes can then bi-orient in a cohesin-independent manner to restrain untimely spindle extension.
Figures







References
-
- Agarwal, R., Z. Tang, H. Yu, and O. Cohen-Fix. 2003. Two distinct pathways for inhibiting Pds1 ubiquitination in response to DNA damage. J. Biol. Chem. 278:45027–45033. - PubMed
-
- Alcasabas, A., A. Osborn, J. Bachant, F. Hu, P. Werler, K. Bousset, K. Kanji-Furuya, J.F.X. Diffley, A. Carr, and S.J. Elledge. 2001. Mrc1 transduces DNA replication stress signals to activate Rad53. Nat. Cell Biol. 3:958–965. - PubMed
-
- Allen, J.B., Z. Zhou, W. Siede, E.C. Friedberg, and S.J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401–2415. - PubMed
-
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S.J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 9:1169–1182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

