Transcriptional regulation by the numbers: models
- PMID: 15797194
- PMCID: PMC3482385
- DOI: 10.1016/j.gde.2005.02.007
Transcriptional regulation by the numbers: models
Abstract
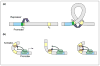
The expression of genes is regularly characterized with respect to how much, how fast, when and where. Such quantitative data demands quantitative models. Thermodynamic models are based on the assumption that the level of gene expression is proportional to the equilibrium probability that RNA polymerase (RNAP) is bound to the promoter of interest. Statistical mechanics provides a framework for computing these probabilities. Within this framework, interactions of activators, repressors, helper molecules and RNAP are described by a single function, the "regulation factor". This analysis culminates in an expression for the probability of RNA polymerase binding at the promoter of interest as a function of the number of regulatory proteins in the cell.
Figures


References
-
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R. Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev. 2005;15:125–135. The companion paper to this article applies the thermodynamic models to a host of different promoters in bacteria and shows the regulation factor in action. - PMC - PubMed
-
- Ptashne M. A Genetic Switch. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2004. This book is a reprinting of Ptashne’s classic, with a special additional chapter that examines recent developments concerning regulation of the life cycle of phage lambda. One of the key recent developments is an appreciation of the role of DNA looping in this system.
-
- Ptashne M, Gann A. Genes and Signals. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2002.
-
- Bellomy GR, Mossing MC, Record MT. Physical properties of DNA in vivo as probed by the length dependence of the lac operator looping process. Biochemistry. 1988;27:3900–3906. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

