Identification of a novel, intraperoxisomal pex14-binding site in pex13: association of pex13 with the docking complex is essential for peroxisomal matrix protein import
- PMID: 15798189
- PMCID: PMC1069607
- DOI: 10.1128/MCB.25.8.3007-3018.2005
Identification of a novel, intraperoxisomal pex14-binding site in pex13: association of pex13 with the docking complex is essential for peroxisomal matrix protein import
Abstract
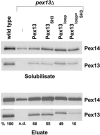
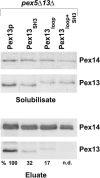
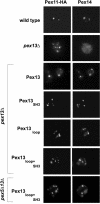
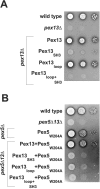
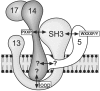
The peroxisomal docking complex is a key component of the import machinery for matrix proteins. The core protein of this complex, Pex14, is thought to represent the initial docking site for the import receptors Pex5 and Pex7. Associated with this complex is a fraction of Pex13, another essential component of the import machinery. Here we demonstrate that Pex13 directly binds Pex14 not only via its SH3 domain but also via a novel intraperoxisomal site. Furthermore, we demonstrate that Pex5 also contributes to the association of Pex13 with Pex14. Peroxisome function was affected only mildly by mutations within the novel Pex14 interaction site of Pex13 or by the non-Pex13-interacting mutant Pex5(W204A). However, when these constructs were tested in combination, PTS1-dependent import and growth on oleic acid were severely compromised. When the SH3 domain-mediated interaction of Pex13 with Pex14 was blocked on top of that, PTS2-dependent matrix protein import was completely compromised and Pex13 was no longer copurified with the docking complex. We conclude that the association of Pex13 with Pex14 is an essential step in peroxisomal protein import that is enabled by two direct interactions and by one that is mediated by Pex5, a result which indicates a novel, receptor-independent function of Pex5.
Figures



Similar articles
-
The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes.Plant J. 2006 Aug;47(4):604-18. doi: 10.1111/j.1365-313X.2006.02809.x. Epub 2006 Jun 30. Plant J. 2006. PMID: 16813573
-
Saccharomyces cerevisiae Pex14p contains two independent Pex5p binding sites, which are both essential for PTS1 protein import.FEBS Lett. 2005 Jun 20;579(16):3416-20. doi: 10.1016/j.febslet.2005.05.011. FEBS Lett. 2005. PMID: 15949802
-
Functional analysis of PEX13 mutation in a Zellweger syndrome spectrum patient reveals novel homooligomerization of PEX13 and its role in human peroxisome biogenesis.Hum Mol Genet. 2013 Oct 1;22(19):3844-57. doi: 10.1093/hmg/ddt238. Epub 2013 May 27. Hum Mol Genet. 2013. PMID: 23716570
-
Opinion: peroxisomal-protein import: is it really that complex?Nat Rev Mol Cell Biol. 2002 May;3(5):382-9. doi: 10.1038/nrm807. Nat Rev Mol Cell Biol. 2002. PMID: 11988772 Review.
-
The peroxisomal protein import machinery.FEBS Lett. 2007 Jun 19;581(15):2811-9. doi: 10.1016/j.febslet.2007.04.001. Epub 2007 Apr 9. FEBS Lett. 2007. PMID: 17445803 Review.
Cited by
-
Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat--store 'em up or burn 'em down.Genetics. 2013 Jan;193(1):1-50. doi: 10.1534/genetics.112.143362. Genetics. 2013. PMID: 23275493 Free PMC article. Review.
-
Glycosome heterogeneity in kinetoplastids.Biochem Soc Trans. 2021 Feb 26;49(1):29-39. doi: 10.1042/BST20190517. Biochem Soc Trans. 2021. PMID: 33439256 Free PMC article. Review.
-
A defect in the peroxisomal biogenesis in germ cells induces a spermatogenic arrest at the round spermatid stage in mice.Sci Rep. 2019 Jul 2;9(1):9553. doi: 10.1038/s41598-019-45991-6. Sci Rep. 2019. PMID: 31267012 Free PMC article.
-
PEX5 and ubiquitin dynamics on mammalian peroxisome membranes.PLoS Comput Biol. 2014 Jan;10(1):e1003426. doi: 10.1371/journal.pcbi.1003426. Epub 2014 Jan 16. PLoS Comput Biol. 2014. PMID: 24453954 Free PMC article.
-
PEX14 binding to Arabidopsis PEX5 has differential effects on PTS1 and PTS2 cargo occupancy of the receptor.FEBS Lett. 2014 Jun 27;588(14):2223-9. doi: 10.1016/j.febslet.2014.05.038. Epub 2014 May 28. FEBS Lett. 2014. PMID: 24879895 Free PMC article.
References
-
- Agne, B., N. M. Meindl, K. Niederhoff, H. Einwachter, P. Rehling, A. Sickmann, H. E. Meyer, W. Girzalsky, and W. H. Kunau. 2003. Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11:635-646. - PubMed
-
- Albertini, M., P. Rehling, R. Erdmann, W. Girzalsky, J. A. Kiel, M. Veenhuis, and W. H. Kunau. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89:83-92. - PubMed
-
- Blobel, F., and R. Erdmann. 1996. Identification of a yeast peroxisomal member of the family of AMP-binding proteins. Eur. J. Biochem. 240:468-476. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
