Histone dynamics on the interleukin-2 gene in response to T-cell activation
- PMID: 15798206
- PMCID: PMC1069623
- DOI: 10.1128/MCB.25.8.3209-3219.2005
Histone dynamics on the interleukin-2 gene in response to T-cell activation
Abstract
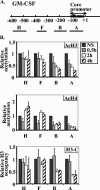
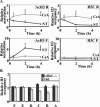
Several models have been proposed for the mechanism of chromatin remodelling across the promoters of inducible genes in mammalian cells. The most commonly held model is one of cooccupation where histone proteins are modified by acetylation or phosphorylation and nucleosomes are remodelled, allowing the assembly of transcription factor complexes. Using chromatin immunoprecipitation, we observed an apparent decrease of histone acetylation and phosphorylation signals at the proximal promoter region of the inducible interleukin-2 and granulocyte-macrophage colony-stimulating factor genes in response to T-cell activation. We showed that this apparent decrease was due to a loss of histone H3 and H4 proteins corresponding to a decrease in nucleosome occupation of the promoter. This histone loss is reversible; it is dependent on the continual presence of appropriate activating signals and transcription factors and is not dependent on the acetylation status of the histone proteins. These data show for the first time that histone proteins are lost from a mammalian promoter upon activation of transcription and support a model of activation-dependent disassembly and reassembly of nucleosomes.
Figures






Similar articles
-
T-cell specific enhancement of histone H3 acetylation in 5' flanking region of the IL-2 gene.Biochem Biophys Res Commun. 2005 Jun 3;331(2):589-94. doi: 10.1016/j.bbrc.2005.03.216. Biochem Biophys Res Commun. 2005. PMID: 15850801
-
H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes.Mol Microbiol. 2006 Dec;62(5):1433-46. doi: 10.1111/j.1365-2958.2006.05451.x. Mol Microbiol. 2006. PMID: 17121596
-
A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly.EMBO J. 2007 Jun 20;26(12):2868-79. doi: 10.1038/sj.emboj.7601728. Epub 2007 May 17. EMBO J. 2007. PMID: 17510629 Free PMC article.
-
Inducible covalent posttranslational modification of histone H3.Sci STKE. 2005 Apr 26;2005(281):re4. doi: 10.1126/stke.2812005re4. Sci STKE. 2005. PMID: 15855410 Review.
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
Cited by
-
Roles of the NF-kappaB pathway in lymphocyte development and function.Cold Spring Harb Perspect Biol. 2010 May;2(5):a000182. doi: 10.1101/cshperspect.a000182. Epub 2009 Dec 23. Cold Spring Harb Perspect Biol. 2010. PMID: 20452952 Free PMC article. Review.
-
Differentiation-dependent functional and epigenetic landscapes for cytokine genes in virus-specific CD8+ T cells.Proc Natl Acad Sci U S A. 2011 Sep 13;108(37):15306-11. doi: 10.1073/pnas.1112520108. Epub 2011 Aug 29. Proc Natl Acad Sci U S A. 2011. PMID: 21876173 Free PMC article.
-
Inducible nucleosome depletion at OREBP-binding-sites by hypertonic stress.PLoS One. 2009 Dec 24;4(12):e8435. doi: 10.1371/journal.pone.0008435. PLoS One. 2009. PMID: 20041176 Free PMC article.
-
Molecular dissection of formation of senescence-associated heterochromatin foci.Mol Cell Biol. 2007 Mar;27(6):2343-58. doi: 10.1128/MCB.02019-06. Epub 2007 Jan 22. Mol Cell Biol. 2007. PMID: 17242207 Free PMC article.
-
Immunopathogenic interaction of environmental triggers and genetic susceptibility in diabetes: is epigenetics the missing link?Diabetes. 2008 Dec;57(12):3184-6. doi: 10.2337/db08-1275. Diabetes. 2008. PMID: 19033405 Free PMC article. No abstract available.
References
-
- Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. - PubMed
-
- Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. - PubMed
-
- Arney, K. L., and A. G. Fisher. 2004. Epigenetic aspects of differentiation. J. Cell Sci. 117:4355-4363. - PubMed
-
- Attema, J. L., R. Reeves, V. Murray, I. Levichkin, M. D. Temple, D. J. Tremethick, and M. F. Shannon. 2002. The human IL-2 gene promoter can assemble a positioned nucleosome that becomes remodeled upon T cell activation. J. Immunol. 169:2466-2476. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
