Floral gene resources from basal angiosperms for comparative genomics research
- PMID: 15799777
- PMCID: PMC1083416
- DOI: 10.1186/1471-2229-5-5
Floral gene resources from basal angiosperms for comparative genomics research
Abstract
Background: The Floral Genome Project was initiated to bridge the genomic gap between the most broadly studied plant model systems. Arabidopsis and rice, although now completely sequenced and under intensive comparative genomic investigation, are separated by at least 125 million years of evolutionary time, and cannot in isolation provide a comprehensive perspective on structural and functional aspects of flowering plant genome dynamics. Here we discuss new genomic resources available to the scientific community, comprising cDNA libraries and Expressed Sequence Tag (EST) sequences for a suite of phylogenetically basal angiosperms specifically selected to bridge the evolutionary gaps between model plants and provide insights into gene content and genome structure in the earliest flowering plants.
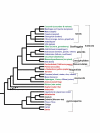
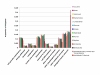
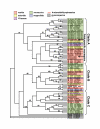
Results: Random sequencing of cDNAs from representatives of phylogenetically important eudicot, non-grass monocot, and gymnosperm lineages has so far (as of 12/1/04) generated 70,514 ESTs and 48,170 assembled unigenes. Efficient sorting of EST sequences into putative gene families based on whole Arabidopsis/rice proteome comparison has permitted ready identification of cDNA clones for finished sequencing. Preliminarily, (i) proportions of functional categories among sequenced floral genes seem representative of the entire Arabidopsis transcriptome, (ii) many known floral gene homologues have been captured, and (iii) phylogenetic analyses of ESTs are providing new insights into the process of gene family evolution in relation to the origin and diversification of the angiosperms.
Conclusion: Initial comparisons illustrate the utility of the EST data sets toward discovery of the basic floral transcriptome. These first findings also afford the opportunity to address a number of conspicuous evolutionary genomic questions, including reproductive organ transcriptome overlap between angiosperms and gymnosperms, genome-wide duplication history, lineage-specific gene duplication and functional divergence, and analyses of adaptive molecular evolution. Since not all genes in the floral transcriptome will be associated with flowering, these EST resources will also be of interest to plant scientists working on other functions, such as photosynthesis, signal transduction, and metabolic pathways.
Figures

References
-
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. - DOI - PubMed
-
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

