Analysis of host response to bacterial infection using error model based gene expression microarray experiments
- PMID: 15800204
- PMCID: PMC1072804
- DOI: 10.1093/nar/gni050
Analysis of host response to bacterial infection using error model based gene expression microarray experiments
Erratum in
- Nucleic Acids Res. 2005;33(7):2352-3
Abstract
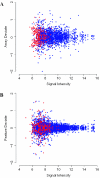
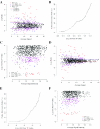
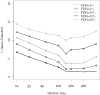
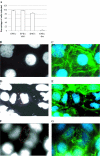
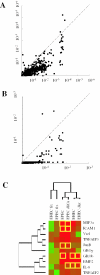
A key step in the analysis of microarray data is the selection of genes that are differentially expressed. Ideally, such experiments should be properly replicated in order to infer both technical and biological variability, and the data should be subjected to rigorous hypothesis tests to identify the differentially expressed genes. However, in microarray experiments involving the analysis of very large numbers of biological samples, replication is not always practical. Therefore, there is a need for a method to select differentially expressed genes in a rational way from insufficiently replicated data. In this paper, we describe a simple method that uses bootstrapping to generate an error model from a replicated pilot study that can be used to identify differentially expressed genes in subsequent large-scale studies on the same platform, but in which there may be no replicated arrays. The method builds a stratified error model that includes array-to-array variability, feature-to-feature variability and the dependence of error on signal intensity. We apply this model to the characterization of the host response in a model of bacterial infection of human intestinal epithelial cells. We demonstrate the effectiveness of error model based microarray experiments and propose this as a general strategy for a microarray-based screening of large collections of biological samples.
Figures
Similar articles
-
Cross platform microarray analysis for robust identification of differentially expressed genes.BMC Bioinformatics. 2007 Mar 8;8 Suppl 1(Suppl 1):S5. doi: 10.1186/1471-2105-8-S1-S5. BMC Bioinformatics. 2007. PMID: 17430572 Free PMC article.
-
A mixture model with random-effects components for clustering correlated gene-expression profiles.Bioinformatics. 2006 Jul 15;22(14):1745-52. doi: 10.1093/bioinformatics/btl165. Epub 2006 May 3. Bioinformatics. 2006. PMID: 16675467
-
EVE (external variance estimation) increases statistical power for detecting differentially expressed genes.Plant J. 2007 Nov;52(3):561-9. doi: 10.1111/j.1365-313X.2007.03227.x. Epub 2007 Aug 3. Plant J. 2007. PMID: 17680783
-
Long versus short oligonucleotide microarrays for the study of gene expression in nonhuman primates.J Neurosci Methods. 2006 Apr 15;152(1-2):179-89. doi: 10.1016/j.jneumeth.2005.09.007. Epub 2005 Oct 25. J Neurosci Methods. 2006. PMID: 16253343
-
Statistical issues in the design and analysis of gene expression microarray studies of animal models.J Mammary Gland Biol Neoplasia. 2003 Jul;8(3):359-74. doi: 10.1023/b:jomg.0000010035.57912.5a. J Mammary Gland Biol Neoplasia. 2003. PMID: 14973379 Review.
Cited by
-
Deep analysis of cellular transcriptomes - LongSAGE versus classic MPSS.BMC Genomics. 2007 Sep 24;8:333. doi: 10.1186/1471-2164-8-333. BMC Genomics. 2007. PMID: 17892551 Free PMC article.
-
Transcriptome Analysis Reveals Modulation of Human Stem Cells from the Apical Papilla by Species Associated with Dental Root Canal Infection.Int J Mol Sci. 2022 Nov 20;23(22):14420. doi: 10.3390/ijms232214420. Int J Mol Sci. 2022. PMID: 36430898 Free PMC article.
-
Shiga toxin remodels the intestinal epithelial transcriptional response to Enterohemorrhagic Escherichia coli.PLoS Pathog. 2021 Feb 2;17(2):e1009290. doi: 10.1371/journal.ppat.1009290. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33529199 Free PMC article.
-
A Network Biology Approach Identifies Molecular Cross-Talk between Normal Prostate Epithelial and Prostate Carcinoma Cells.PLoS Comput Biol. 2016 Apr 28;12(4):e1004884. doi: 10.1371/journal.pcbi.1004884. eCollection 2016 Apr. PLoS Comput Biol. 2016. PMID: 27124473 Free PMC article.
-
Genome-wide identification of Chlamydia trachomatis antigens associated with trachomatous trichiasis.Invest Ophthalmol Vis Sci. 2012 May 14;53(6):2551-9. doi: 10.1167/iovs.11-9212. Print 2012 May. Invest Ophthalmol Vis Sci. 2012. PMID: 22427578 Free PMC article.
References
-
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Chin K.V., Kong A.N. Application of DNA microarrays in pharmacogenomics and toxicogenomics. Pharm. Res. 2002;19:1773–1778. - PubMed
-
- Butte A. The use and analysis of microarray data. Nature Rev. Drug. Discov. 2002;1:951–960. - PubMed
-
- Heller M.J. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4:129–153. - PubMed
-
- Shirota Y., Kaneko S., Honda M., Kawai H.F., Kobayashi K. Identification of differentially expressed genes in hepatocellular carcinoma with cDNA microarrays. Hepatology. 2001;33:832–840. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

